More Information
Submitted: November 01, 2023 | Approved: November 20, 2023 | Published: November 21, 2023
How to cite this article: Otun S, Achilonu I. Exploring the Complexity of Protein Structure Determination Through X-ray Diffraction. J Plant Sci Phytopathol. 2023; 7: 124-132.
DOI: 10.29328/journal.jpsp.1001117
Copyright License: © 2023 Otun S, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Exploring the Complexity of Protein Structure Determination Through X-ray Diffraction
Sarah Otun* and Ikechukwu Achilonu
Department of Molecular and Cell Biology, University of the Witwatersrand, South Africa
*Address for Correspondence: Sarah Otun, Department of Molecular and Cell Biology, University of the Witwatersrand, South Africa, Email: [email protected]
The determination of a protein structure by using X-ray diffraction encompasses a series of sequential steps (including gene identification and cloning, protein expression and purification, crystallization, phasing model building, refinement, and validation), which need the application of several methodologies derived from molecular biology, bioinformatics, and physical sciences. This article thoroughly examines the complicated procedure of elucidating protein structures within plant biology, using X-ray diffraction as the primary methodology. Commencing with the gene identification process and progressing toward crystallography, this article explores the many obstacles and achievements in acquiring diffraction pictures and their subsequent conversion into electron density maps. The ensuing phases of model construction, refinement, and structural validation are thoroughly examined, providing insight into the inherent complexity associated with each stage. The paper also discusses the critical component of understanding the resultant model and scrutinizing its biological significance. By comprehensively examining these stages, this article presents a nuanced comprehension of the intricate procedure in ascertaining protein structures within plant biology. It offers valuable insights into the obstacles encountered and the biological importance of the acquired structural data.
Protein crystallography, along with the broader field of structural biology, has greatly benefited from several remarkable research throughout its historical development and continues to do so today. A notable example is Max Perutz's extensive investigation into the structure of hemoglobin for a duration exceeding 20 years before the publication of the first noteworthy findings in 1960 [1]. Nonetheless, his contributions facilitated the emergence of a novel approach that subsequently gained widespread adoption among other research groups exploring protein structures. Consequently, in the year 1962, Perutz was honoured with the prestigious Nobel Prize in chemistry [2]. Furthermore, the selection of haemoglobin as the focal point of this endeavour proved advantageous due to its extremely high proportion of a-helical secondary structure. This characteristic imparts significant rigidity, stability, and favourable diffraction properties to the protein, making it very straightforward to model.
Subsequently, the advent of synchrotrons by researchers in the field of high-energy physics had a pivotal role in facilitating the rapid increase in the number of protein structures that were successfully determined using X-ray radiation. The particles in orbit, namely electrons or positrons, produce a kind of radiation formerly referred to as parasitic radiation during the early stages. Hence, a minute aperture in the synchrotron enclosure can provide an x-radiation of much greater intensity than conventional generators. Since the early 1980s, several synchrotron X-ray sources have been constructed globally. These first installations were succeeded by third-generation facilities, which produce x-rays by the conventional circulation of particles around the rings and use specialized insertion devices known as wigglers and undulators.
Crystallographers possess over 100 specialised X-ray beamlines on 22 synchrotrons established throughout all continents except Antarctica. In 2005, synchrotron sources determined 3897 protein structures, accounting for about 75% of the total protein structures reported. After placing crystals in the synchrotron beam, several structures were solved over a very short period, ranging from hours to minutes [3].
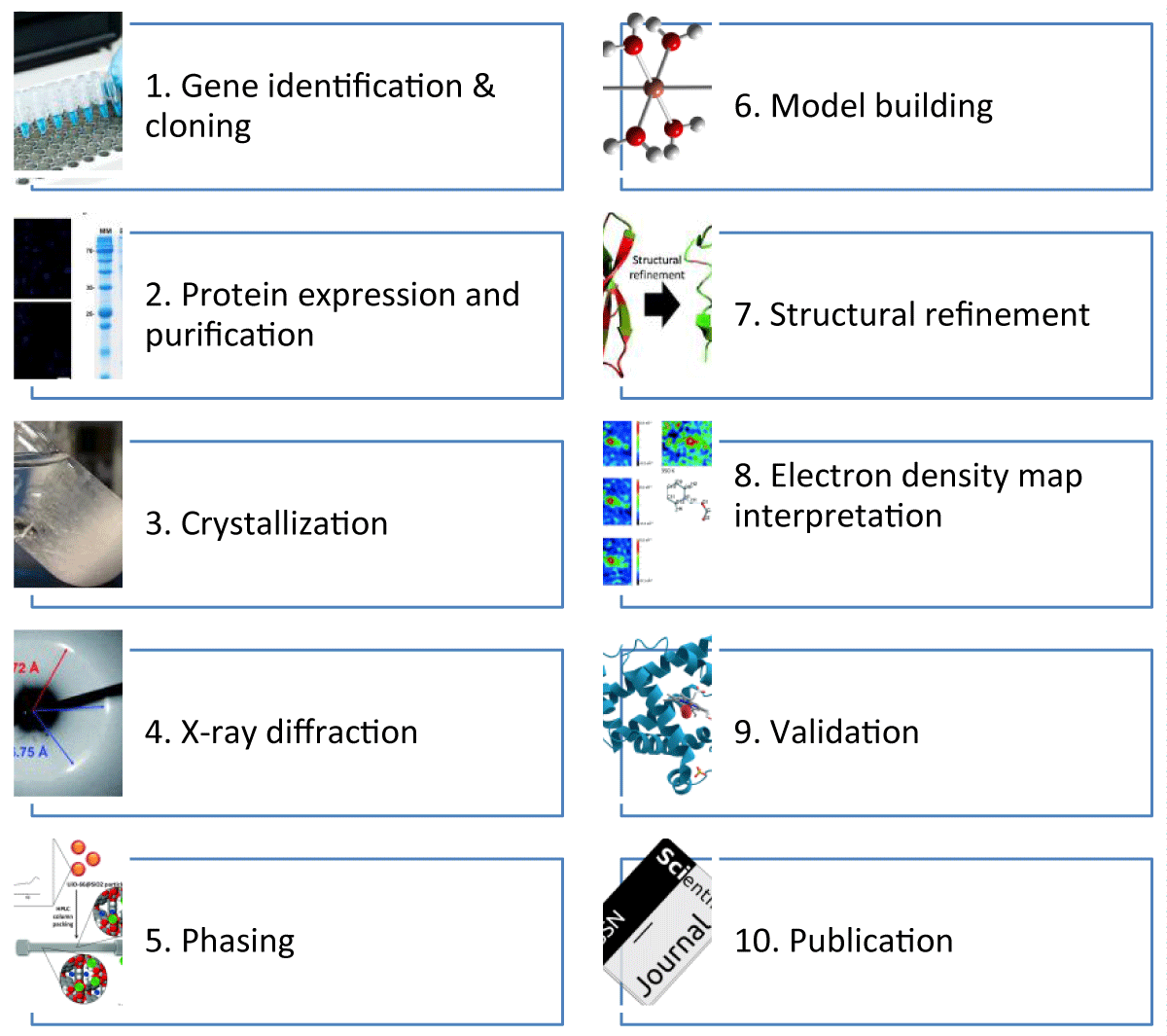
The progress in X-ray sources has been paralleled by the emergence of rapid X-ray detectors and significant advancements in computational techniques and computer technology. These developments happened simultaneously when molecular biology techniques were undergoing rapid expansion. Affordable workstations or laptops have sufficient computing capabilities to resolve most crystallographic structures. Moreover, highly advanced software can effectively determine the three-dimensional arrangement, even when crystals with low diffraction quality are used. The method of determining the structure of a macromolecule involves many steps, as shown in Figure 1. Each step must be completed to explain the macromolecule's structure comprehensively. One of the primary challenges encountered in protein crystallography is the assessment of the efficacy of a given phase, which can only be thoroughly examined at subsequent stages and, in some cases, even after two or three following steps. The researcher may need to revisit a prior (or the first) stage to get optimal outcomes. The process of structural solution via iteration may require a significant time commitment, perhaps spanning a period of 10 to 20 years, as researchers engage in multifaceted efforts.
Figure 1: Schematic Overview of the X-ray Crystallography Workflow.
The figure also represents the step-by-step process from gene identification to the publication of the protein structure.
Notable examples include the determination of the structure of nerve growth factor, which was solved 17 years after the availability of crystals [4] and the structure of L-asparaginase, which was solved 19 years after the first crystallization [5]. This review aims to analyse the challenges associated with the multiple stages involved in the progression from a gene to the ultimate publication, encompassing elucidating a macromolecule's structure and mechanism of action. Additionally, it will explore the advantageous or adverse circumstances that may impact the pace of progress along this trajectory.
Transforming gene to crystal structure
In a high-throughput structural genomics centre, the first stage is a comprehensive examination of all accessible data about a protein target using bioinformatic and experimental methodologies. In many instances, the empirical understanding of a protein may be lacking or limited. Nevertheless, several bioinformatic methods can derive valuable insights, even when the only accessible information pertains to the gene sequence. For instance, these methods often facilitate the removal of inherently unstructured proteins during the first phases of the procedure. There is a consensus among researchers that studying mammalian and membrane proteins presents more challenges compared to soluble bacterial proteins. However, it is essential to note that even a seemingly simple bacterial protein may pose significant difficulties and require much effort to determine its structure. Projects exploring the structural aspects of protein complexes may provide an even more incredible problem than research focused on integral membrane proteins. Although chance may play a significant role, solving such structures can still take many years. For example, the process of crystallising ribosomal particles was initiated in 1982 [6]. However, it was not until 2000 that the first comprehensive structures were successfully determined [7].
The first step in determining the protein structure via X-ray diffraction is the cloning process, aided by a wide range of commercially accessible kits and services, such as de novo gene synthesis, simplifying the procedure [8]. Synthetic genes containing optimised codons can enhance protein production, mainly when these genes are produced in a system that employs codon frequencies distinct from those found in their original genome [9]. Upon first examination, the measure of success for this stage is straightforward since it can be determined if the gene has been cloned effectively or not. Regrettably, it is not uncommon for subsequent issues to arise, such as inadequate expression levels, insufficient solubility of proteins, unsuccessful crystallisation attempts, or problematic characteristics of the crystals (e.g., twinning, low-resolution diffraction) [10]. Consequently, it may be necessary to repeat the cloning process, even if it initially appeared successful.
Hence, it is essential to design constructs under the premise that the protein itself is a crucial determinant influencing crystallisation and that the investigation of its structure may require substantial quantities of protein [11]. Selecting a suitable expression system and vector is worthwhile because of its significant impact on research outcomes. Determining whether a protein exhibits autonomous folding or requires specific environmental conditions for optimal folding might have implications for vector design [12]. For example, the target protein may necessitate the presence of an adjunct protein that functions as a molecular chaperone, shielding it from degradation during the process of expression or aiding in the establishment of disulfide bonds [13]. Crystallisation studies can take time, ranging from weeks to months, necessitating the stability of the protein over extended durations. Posttranslational modifications are often seen in proteins derived from eukaryotic organisms [14]. In some instances, the production of a protein in a bacterial environment may provide an inactive form. However, this seemingly unsuccessful outcome might still be helpful for crystallisation, particularly in circumstances where the protein lacks glycosylation. The expression of proteins that include disulfide bonds presents a challenge in achieving the desired folded conformation when using bacterial systems [15]. However, significant attention has been devoted to advancing innovative refolding methodologies owing to the purifying capabilities of some proteins that are only synthesized in inclusion bodies. These proteins may be purified under denaturing conditions and refolded. When refolding proves unsuccessful, the only recourse is to choose an alternative expression protocol or expression system [16].
In some cases, the researcher may be compelled to alter their approach and choose the selection of a fragment as the target for structure determination due to the challenges encountered in purifying and crystallising a whole protein. Limited proteolysis is the preferred method for establishing the borders of domains in multidomain proteins [17]. However, selecting a protein fragment that is both stable and accurately represents a single domain might pose challenges. Also, the expression of mammalian proteins in bacterial hosts might present difficulties, necessitating alternative expression methods such as yeast, insect, or mammalian cells. Throughout each of these steps, it is essential to consider the ultimate objective of the experiment, which is to get a comprehensive understanding of a three-dimensional structure that accurately reflects a physiologically significant shape.
After determining that the protein expression yield is satisfactory, the subsequent step involves the purification of the protein. One century ago, scientists used the crystallisation technique for protein purification; however, in contemporary times, there is a prevailing assumption that the sample must possess homogeneity to achieve the crystallisation of recalcitrant proteins [18]. This homogeneity encompasses the polypeptide sequence, protein folding, conformation, and perhaps aggregation state [19]. To enhance the efficiency and expediency of protein purification, it is common practice to merge recombinant proteins with polypeptides or whole protein partners, facilitating affinity chromatography. The purification procedure that is widely used is the incorporation of a poly-histidine tag (Histag) in conjunction with metal-ion affinity chromatography [20]. This tag comprises 6-10 histidine residues, often accompanied by a spacer that facilitates subsequent cleavage by the action of an appropriate protease. One notable benefit of the His-tag is its compact nature, which sometimes obviates the need for tag removal before crystallization [21]. Incorporating fusion proteins often enhances the magnitude of protein production and its solubility. The fusion of maltose-binding protein, thioredoxin, glutathione-S-transferase, or green fluorescent protein has been shown to provide many potential benefits in several scenarios. These proteins may be used individually or with a His-tag. Thus, including a fusion partner, namely maltose-binding protein, has been shown to facilitate the correct folding of the passenger protein [22].
Furthermore, the crystallisation process necessitates the availability of protein samples in several milligrams. Nevertheless, the progress made in nanotechnologies and serendipitous events may significantly decrease the amount of protein required to provide the first conditions for crystallisation. Regrettably, nanoliter technologies often provide crystals of insufficient size for X-ray structural research. Scaling up nanoliter crystallisation conditions to the microliter scale is sometimes challenging and yet to be understood entirely. However, using on-chip methods has promise in addressing this knowledge gap [23]. The primary approach for initial screening is often the sparse matrix technique [24]. Many commercially available screens have been specifically designed to optimise the crystallisation of proteins, nucleic acids, protein complexes, and membrane proteins [25]. If the researcher is fortunate, upon establishing several crystallisation conditions, they may proceed with the optimisation of crystal development. Crystal optimisation may be carried out using several methods, with grid screen designs often used in most procedures, depending on the first acquired circumstances [26].
Additional methods include incorporating additives, often called small-molecule compounds, into the crystallisation medium [27]. Researchers who have reported adverse outcomes in their attempts to identify suitable circumstances for crystal formation may consider using reductive methylation of lysine residues as an alternative approach before embarking on developing novel protein structures [28]. When reductive methylation is unsuccessful, the subsequent option for enhancing the crystallizability of a protein might include introducing sequence mutations [29]. However, this strategy necessitates returning to the process's first stage.
One alternative rescue operation approach involves using in situ proteolysis [30]. Many diffraction experiments are currently conducted in synchrotron facilities. The flux at some beamlines with high intensity is of such magnitude that an unshielded crystal would undergo evaporation within a matter of milliseconds. Synchrotron beams of very moderate strength have been seen to elicit radiation damage, which in turn may lead to a range of chemical changes occurring inside the protein [31].
Moreover, Cryocooling has emerged as the most effective approach for decelerating the abovementioned process [32]. When combined with the straightforward cryo-loop crystal mounting method, this technique has brought about a significant transformation in data collecting [33]. During Cryocooling, crystals that cryo solutions have safeguarded are expeditiously moved to a nitrogen stream that is consistently kept at a temperature near 100 Kelvin. In such circumstances, the solution inside and around the crystal undergoes vitrification. The cryosolutions may consist of various alcohols, salts, or oils that inhibit ice formation, preserving protein crystals' structural integrity. Although conceptually straightforward, the process of crystal freezing requires the evaluation of many cryosolutions. However, even comprehensive cryocooling investigations may yield specimens of significantly diminished quality compared to the initial crystals. The manipulation of the crystal environment, mainly via the regulation of humidity or through the process of annealing, can significantly enhance the quality of crystals.
The next step is to generate a density map—a topographic map with contour lines at intervals of 1 unit. The crystal was acquired by utilizing naturally occurring protein derived from soybeans, subsequently identifying the residue as serine (PDB codes: 1YGE and 1F8N). The map is shown at a time interval of 0.7 seconds. The replacement of serine with glutamic acid is consistent with the outcomes of the DNA sequencing analysis, as Ted Holman reported in a private correspondence in 2007. The electron density is shown at a time interval of 0.3 seconds. Various contouring techniques on the map may provide insights into two potential phenomena: decarboxylation during data gathering or conformational flexibility in Glu160.
Obtaining an electron density map via diffraction images
Positioning a crystal inside the X-ray beam initiates the last stage of the structure determination procedure. The experiment is fundamentally essential, leading to the expectation that automating the data-collecting process should be relatively straightforward. This is due to the limited number of factors the investigator controls. The parameters included in this study are the distance between the crystal and detector, the duration of exposure, the angle of oscillation, and the wavelength of x-radiation [34]. Nevertheless, there exist a minimum of three supplementary variables that fall beyond the jurisdiction of the experimenter: the quality of the crystal (specifically, its long-range order and mosaic spread), radiation decay, and the constraints imposed by the experimental apparatus (such as the dynamic range of the detector and the accuracy of the goniostat, among others). The challenge of selecting user-controlled settings that effectively mitigate the negative impact of crystal quality and radiation decay is shown by examining the data obtained from one of the beamlines at the Advanced Light Source (ALS). The results obtained from the beamline indicate that, on average, a total of 57 complete data sets must be collected to successfully make a Protein Results Bank (PDB) deposit [35]. Furthermore, it should be noted that the number of crystals evaluated is significantly greater. The experimental challenge arises from the observation that a diffraction experiment yields a collection of diffraction intensities (or amplitudes) rather than the phases required to compute the electron density map.
In contemporary scientific methodology, the acquisition of diffraction data is undertaken for three primary computational purposes: molecular replacement (MR), multiple anomalous diffraction (MAD)/single anomalous diffraction (SAD), and the ultimate refining of the model [36]. The method of multiple isomorphous replacements, which was formerly widely favoured, has been surpassed in popularity by procedures that rely on anomalous scattering. In magnetic resonance (MR) experiments, the origin of phases is derived from a model of the same or a comparable protein [37]. In this scenario, the precision of the measured intensities is of lesser significance than acquiring a comprehensive dataset while ensuring that solid peaks are preserved due to detector oversaturation.
Accurately determining phases is crucial for solving novel structures using Single-wavelength Anomalous Dispersion (SAD), Multiple-wavelength Anomalous Dispersion (MAD), or multiple isomorphous replacements [38]. These methods obtain the phases by calculating the differences between the measured diffraction intensities. The primary objective of data gathering for the final refining of the model is to gather comprehensive and highly accurate data that reaches the resolution limit of diffraction. The second experiment is less challenging, but it still needs meticulous preparation. Enhancing the statistical precision of measured intensities does not automatically lead to improved data quality since prolonged counting time may exacerbate radiation damage.
Historically, investigations using SAD/MAD, which need precise data, were notably challenging. However, advancements in experimental gear, software, and procedures have significantly improved the success rate of determining structures using these approaches. Given that a Single Acquisition Diffraction (SAD) experiment entails gathering only a portion of the data necessary for Multiple Acquisition Diffraction (MAD), one would anticipate that the former approach would be favoured. However, the variations observed across different global regions in adopting these methodologies indicate sluggish dissemination of the most compelling experimental protocols. Experiments utilizing Single-wavelength Anomalous Dispersion (SAD) or Multiple-wavelength Anomalous Dispersion (MAD) techniques seldom encounter failure solely attributable to insufficient atoms generating an anomalous signal [39]. However, the primary causes of experimental failure are typically related to errors in the procedure, such as an excessive number of saturated detector pixels (overloads) or an inadequate data collection strategy that may lead to premature radiation damage or incomplete data. It is worth noting that using weak anomalous scatterers, such as sulphur, may present an exception to this observation.
Minor mistakes made during this phase result in a substantial increase in the workload for the researcher throughout the processes of structure determination and refining. In several instances, it is necessary to repeat diffraction studies that fail to achieve ideal results. The issue of neglecting the completeness of low-resolution data is a common occurrence, as shown by the fact that the Protein Data Bank (PDB) reports completeness only in the highest-resolution shell and not in the lowest. This oversight may lead to structure-solving and model-building problems [40]. Occasionally, individuals may successfully resolve such issues using unconventional methods by combining fortuitous circumstances and expertise. However, it is essential to emphasize that precise, unsaturated, low-resolution data play a crucial role in MR and SAD/MAD procedures. It might sometimes be unexpected that structural analysis can be resolved using data obtained from a crystal of poorer quality instead of relying on data taken from a crystal of higher quality. Crystals of inferior quality exhibit limited diffraction capabilities, resulting in the generation of low-intensity reflections that do not contribute significantly to the resolution of the crystal structure. In this scenario, the selection sequence of crystals for diffraction tests may impact the likelihood of successful outcomes. It is uncommon for researchers to gather an additional, comprehensive dataset if they have previously seen diffraction patterns that are deemed "perfect."
Over the last several years, the analysis and interpretation of experimental findings have been significantly enhanced by using many integrated software packages, including CCP4, PHENIX, and HKL-3000 [41-43]. This progress has been further supported by the accessibility of high-performance computers, enabling almost instantaneous computations throughout the analysis process. Despite their inherent complexity, the techniques for resolving the phase issue are concealed behind advanced software and, at times, even more advanced user interfaces. These tools enable those needing more crystallography expertise to work effectively and proficiently. A comprehensible initial electron density map may be acquired in straightforward scenarios with a few mouse clicks.
Frequently, the first stages (including electron density maps) derived from SAD/MAD or MR investigations tend to exhibit limited accuracy, posing challenges in their interpretation. Fortunately, several strategies for enhancing phase accuracy have been devised, which, when appropriately implemented, can significantly improve the quality of electron density maps. Solvent-flattening techniques and noncrystallographic symmetry averaging are widely used in the field. Notably, crystals containing a significant amount of solvent frequently exhibit poor diffraction patterns. However, it is essential to acknowledge that the high solvent content might provide advantages by facilitating the generation of a high-quality first map. Noncrystallographic symmetry averaging leads to significant variations in the quality of electron density maps, indicating that several instances of a macromolecule in an asymmetric unit should not be regarded as a mere coincidence.
When data gathering is executed with precision, challenges may arise in the structure solution process due to inherent issues associated with the characteristics of the crystals. Twinning, a phenomenon in which several lattices diffract concurrently, presents one of the most challenging challenges. A recent analysis (50) has shown that the coexistence of crystal and lattice symmetries can induce twinning in over 30% of the structures documented in the Protein Data Bank [44]. Furthermore, twinning is not universally discernible, and under some circumstances, it hinders determining the crystal structure. In such an occurrence, the only viable course of action would be to revisit the laboratory setting to cultivate an alternative crystal structure for a particular macromolecule.
Model building, refinement and validation
In contemporary scientific practice, generating initial electron density maps is often facilitated by automated or semi-automated software. Furthermore, the subsequent interpretation of the resultant electron density may also be conducted with a high degree of automation. Various software programs, such as ARP/wARP, RESOLVE, and MAIN, use diverse methodologies for automated model construction [45-47]. Manual model development and tweaking are facilitated by the availability of robust graphics software tools like O and COOT [48,49]. Nevertheless, interpreting electron density maps acquired at low resolution remains a nontrivial task. Protein structural refinement is often conducted using software applications such as CNS/CNX/X-PLOR, REFMAC, and SHELXL —2.0 Å resolution [50-52].
The measurement is 2.4 angstroms. Refining data with a high level of precision may be a time-consuming task, primarily because it involves modeling several intricate structural elements. These elements include different conformations of side chains and complex temperature factor models, among others. Structures with shallow resolution (less than 3.2 A˚) are distinct, necessitating meticulous refining and validation procedures. A further challenge in refining emerges when a molecular structure includes components other than amino acids, such as metal ions and tiny molecule compounds. While identifying and refining metal ions inside protein structures may seem relatively simple, it is worth noting that several newly reported structures in the Protein Data Bank (PDB) still exhibit metal ions with very unlikely coordination or geometric characteristics in their metal-binding environments. The computerized model-building process is most effective for amino-acid chains, which may provide extra challenges when dealing with protein-DNA or protein-ligand complexes [53]. The effectiveness of a refinement method is primarily contingent upon the resolution used. Additionally, the chosen resolution dictates the extent to which parameters may be improved and the appropriate approach for handling them.
The process of refinement and manual structural rebuilding or modification should be conducted in conjunction with model validation [54]. As previously said, notable progress in software has facilitated the ability of several individuals who need to be more specialised in crystallography to gather data and effectively determine and enhance X-ray structures without extensive familiarity with the fundamental methodologies involved. In particular instances such as these, using advanced technologies for structural validation becomes imperative. Validation tools should be able to identify significant crystallographic and chemical inaccuracies in models and provide guidance to those without knowledge, hence offering suggestions on how to rectify these mistakes. Several programs within this category are PROCHECK, WHATCHECK, MOLPROBITY, and KING [55-57].
Sometimes, researchers may disregard explicit cautionary indications provided by validation programs, even throughout the Protein Data Bank (PDB) deposition process. This behaviour may be attributed to their conviction that the structure they have obtained is exceptional, leading them to see any deviations from established chemical principles as evidence supporting its distinctiveness. Regrettably, a limited fraction of fortuitous scientists who see groundbreaking chemical phenomena inside their structures will eventually get recognition from the Nobel Committee. Conversely, those less fortunate will inevitably discover that validation instruments undermine their claims at some point. Given the current mandate for depositing structure factors in most publicly funded research, crystallographers now could employ a highly effective validation tool, namely the re-evaluation of structures that raise doubts. Consequently, the likelihood of erroneous refined structures contaminating databases in the future is minimal.
Protein structure model interpretation
It is imperative to consistently remember that the primary objective of a crystallographic experiment, even when conducted within the framework of structural genomics, is not solely the generation of a model comprising atomic coordinates. Instead, its purpose is to offer valuable insights for interpreting chemical and biological data. Nevertheless, it is crucial to understand the models while considering their limits, which may arise from variables such as data resolution, the overall quality of the model (as shown by R/Rfree), and its chemical accuracy. Furthermore, it is essential to emphasise that the ultimate model does not depict an individual molecule but rather is a representation that accounts for the average characteristics of several molecules throughout time and space. High-energy radiation, specifically emitted by intense synchrotron beamlines, can induce chemical alterations in molecules.
Furthermore, it is essential to note that even a model derived from high-resolution data cannot be regarded as entirely devoid of errors [58]. The potential for misunderstanding of electron density increases with lower resolutions, making it easier to erroneously trace a piece of the amino-acid chain in the other direction or, although rarely, generate an entirely wrong model. Furthermore, how numeric values are represented in the atomic coordinates provided in the PDB format, specifically with three digits after the decimal point, can potentially lead to misinterpretation by inexperienced experimenters. Such individuals may mistakenly assume that all digits hold significance and subsequently analyse the structure based on this assumption [56]. When analysing structures deposited in the Protein Data Bank (PDB), it is essential to consider that these models may contain various errors that arise throughout the structure determination process.
Consequently, the interpretation of a three-dimensional structure and any chemical or biological inferences drawn from it are significantly influenced by the quality of the model [59]. To deduce a comprehensive mechanism of an enzyme process, it is essential to understand the hydrogen-bond network inside the macromolecule under investigation. Regrettably, only a few groups of lucky researchers who can ascertain protein structures at very high levels of precision have the privilege of directly seeing hydrogen atoms inside those structures. Most structures in the Protein Data Bank (PDB), around 60%, exhibit resolutions ranging from 1.7 to 2.5 A˚. Interpreting these structures is not straightforward since a single structure might accommodate various chemical or biological response pathways. A translator of poetry encounters a comparable issue that requires resolution. The translation is a complex and nuanced process that may be considered an artistic endeavour. It is possible for a poem that has been translated into a different language to surpass the quality of the original composition potentially. In many instances, reinterpreting a structure is frequently seen as superior to its original form. The transition from determining coordinates to understanding the mechanism of action is a particularly challenging stage.
Biological relevance assessment
After achieving a high-resolution solution for the structure, characterised by low R factors and little deviation of geometric parameters from library values, what level of confidence may be attributed to its depiction of a physiologically significant state of the protein? The inquiry issue has been posed and subsequently addressed on several occasions since the inception of protein crystallography. The problem at hand encompasses one singular difficulty and a minimum of two interconnected challenges. One of the first inquiries, which may be straightforward to address, pertains to the extent of similarity between the protein's structure in the solid state, namely in the crystal, and its structure in solution.
For instance, after examining the configurations of a little spiral-shaped protein known as interleukin-4, which was resolved autonomously at four separate research facilities. Crystallography was used to acquire two structures of this protein, while two further structures were found using NMR. The analysis conducted by the researchers has effectively shown that the disparities seen between these structures may be attributed only to the inherent uncertainties associated with their determination, with NMR exhibiting much more significant uncertainties compared to crystallography. These discrepancies do not indicate any protein abnormalities [60]. Hence, while acknowledging the validity of this worry and recognising the need to address it on a case-by-case basis, the disparities between the solution and solid state of proteins are often minimal, if existent at all.
However, an additional aspect of the inquiry pertains to the significance of the observed structure in elucidating the biological characteristics of the molecular system being investigated, and this matter needs a definitive response. In this analysis, they examined an enzyme and delved into the intricacies of the process it facilitates. The structural composition of the apoenzyme may provide a partial explanation for some stages of the reaction. This is because some active site components can adapt in response to the presence of the substrate, transition state, and product. Furthermore, the precise nature of these adaptations is often difficult to anticipate. The elucidation of the transition state's structure would provide valuable insights. However, direct access to it is precluded because of its instability within the crystallographic timescale. Applying transition state mimics and rapid data acquisition through Laue crystallography can help [61]. However, they do not offer a definitive assurance that the protein's state observed in the crystal can directly elucidate its biologically significant characteristics, as proteins are inherently dynamic entities.
Another illustrative instance of the many challenges faced in ascertaining the biological characteristics of a protein using crystallographic research is shown by the ATP-dependent protease Lon. Despite being discovered over two decades ago and undergoing crystallographic studies for ten years, the Lon enzyme is resistant to crystallization [62]. Nevertheless, after determining Lon's domain structure, its distinct domains have been subjected to crystallisation and subsequent individual analysis. This study produced some unexpected findings. For instance, variations in the structure of the active site were observed in the catalytic domain of Lon when isolated from various bacterial sources. These observed variances were first hypothesised to have a biological significance [63].
Nevertheless, subsequent investigations using crystallography and mutagenesis techniques have presented a contrasting perspective. These studies propose that the structures of the apoenzyme do not exhibit the active site in a biologically significant state. This is due to the high probability that a substrate or reaction product would cause substantial reorganisation of the active site. The need to identify suitable substrates for Lon hinders a comprehensive understanding of its mode of action despite the availability of an atomic-resolution structure for its catalytic domain [64]. The investigation of the biological features of the N-terminal domain of this protein, which is very probable to be associated with substrate binding, presents an even greater level of complexity. The crystal structure of the construct, including slightly more than 100 residues, revealed a unique fold that has not been seen in any previously characterised protein complexes. Consequently, no definitive inferences on binding could be made.
Nevertheless, the PDB deposition of BPP1347, a putative protein derived from Bordetella parapertussis has shown a remarkable degree of topological resemblance despite little sequence similarity [65]. This instance exemplifies a prevalent issue encountered in specific structural genomics-derived structures, precisely the challenge of attributing function to proteins with a novel fold. Interestingly, this predicament is an explicitly stated motivation behind these endeavours. However, even structures obtained through deliberate and focused initiatives may not significantly improve the situation.
In the field of structural biology, the process of transitioning from gene analysis to the publishing of research findings often requires a substantial investment of effort and a considerable degree of uncertainty. In conclusion, this study reveals the complex path that X-ray diffraction has taken in elucidating the structures of plant proteins. Synergistic integration of molecular biology, bioinformatics, and physical sciences is essential at every stage, from identifying genes to obtaining crystal structures. This review gives a guide through the obstacles and the uncertainty of the success of X-ray diffraction studies. Thus, the review is a compass for future research as the field navigates complexity.
- PERUTZ MF, ROSSMANN MG, CULLIS AF, MUIRHEAD H, WILL G, NORTH AC. Structure of haemoglobin: a three-dimensional Fourier synthesis at 5.5-A. resolution, obtained by X-ray analysis. Nature. 1960 Feb 13;185(4711):416-22. doi: 10.1038/185416a0. PMID: 18990801.
- Seeman JI, Restrepo G. The Mutation of the “Nobel Prize in Chemistry” into the “Nobel Prize in Chemistry or Life Sciences”: Several Decades of Transparent and Opaque Evidence of Change within the Nobel Prize Program. Angewandte Chemie. 2020 Feb 17;132(8):2962–81.
- Fetisov GV. X-ray diffraction methods for structural diagnostics of materials: progress and achievements. Physics-Uspekhi. 2020 Jan 1;63(1):2–32.
- Wlodawer A, Hodgson KO, Shooter EM. Crystallization of nerve growth factor from mouse submaxillary glands. Proc Natl Acad Sci U S A. 1975 Mar;72(3):777-9. doi: 10.1073/pnas.72.3.777. PMID: 1055377; PMCID: PMC432402.
- McDonald NQ, Lapatto R, Murray-Rust J, Gunning J, Wlodawer A, Blundell TL. New protein fold revealed by a 2.3-A resolution crystal structure of nerve growth factor. Nature. 1991 Dec 5;354(6352):411-4. doi: 10.1038/354411a0. PMID: 1956407.
- Yonath A, Müssig J, Wittmann HG. Parameters for crystal growth of ribosomal subunits. J Cell Biochem. 1982;19(2):145-55. doi: 10.1002/jcb.240190205. PMID: 7174745.
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000 Aug 11;289(5481):905-20. doi: 10.1126/science.289.5481.905. PMID: 10937989.
- Kim Y, Babnigg G, Jedrzejczak R, Eschenfeldt WH, Li H, Maltseva N, Hatzos-Skintges C, Gu M, Makowska-Grzyska M, Wu R, An H, Chhor G, Joachimiak A. High-throughput protein purification and quality assessment for crystallization. Methods. 2011 Sep;55(1):12-28. doi: 10.1016/j.ymeth.2011.07.010. Epub 2011 Aug 31. PMID: 21907284; PMCID: PMC3690762.
- Komar AA. The Art of Gene Redesign and Recombinant Protein Production: Approaches and Perspectives. In 2016; 161–77.
- Papageorgiou AC, Poudel N, Mattsson J. Protein Structure Analysis and Validation with X-Ray Crystallography. Methods Mol Biol. 2021;2178:377-404. doi: 10.1007/978-1-0716-0775-6_25. PMID: 33128762.
- Kurpiewska K, Borowski T. Seven quick tips for beginners in protein crystallography. Acta Biochim Pol. 2021 Aug 11;68(4):535-546. doi: 10.18388/abp.2020_5589. PMID: 34379378.
- Liu H, Chen Q. Computational protein design with data‐driven approaches: Recent developments and perspectives. WIREs Computational Molecular Science. 2023 May15;13(3).
- Ye H, Wu J, Liang Z, Zhang Y, Huang Z. Protein S-Nitrosation: Biochemistry, Identification, Molecular Mechanisms, and Therapeutic Applications. J Med Chem. 2022 Apr 28;65(8):5902-5925. doi: 10.1021/acs.jmedchem.1c02194. Epub 2022 Apr 12. PMID: 35412827.
- Macek B, Forchhammer K, Hardouin J, Weber-Ban E, Grangeasse C, Mijakovic I. Protein post-translational modifications in bacteria. Nat Rev Microbiol. 2019 Nov;17(11):651-664. doi: 10.1038/s41579-019-0243-0. Epub 2019 Sep 4. PMID: 31485032.
- Ma Y, Lee CJ, Park JS. Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds. Antibiotics (Basel). 2020 Aug 26;9(9):541. doi: 10.3390/antibiotics9090541. PMID: 32858882; PMCID: PMC7558204.
- Yamaguchi H, Miyazaki M. Refolding techniques for recovering biologically active recombinant proteins from inclusion bodies. Biomolecules. 2014 Feb 20;4(1):235-51. doi: 10.3390/biom4010235. PMID: 24970214; PMCID: PMC4030991.
- Fontana A, de Laureto PP, Spolaore B, Frare E, Picotti P, Zambonin M. Probing protein structure by limited proteolysis. Acta Biochim Pol. 2004;51(2):299-321. PMID: 15218531.
- Giegé R. A historical perspective on protein crystallization from 1840 to the present day. FEBS J. 2013 Dec;280(24):6456-97. doi: 10.1111/febs.12580. Epub 2013 Nov 25. PMID: 24165393.
- Jahn TR, Radford SE. Folding versus aggregation: polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008 Jan 1;469(1):100-17. doi: 10.1016/j.abb.2007.05.015. Epub 2007 Jun 8. PMID: 17588526; PMCID: PMC2706318.
- Mishra V. Affinity Tags for Protein Purification. Curr Protein Pept Sci. 2020;21(8):821-830. doi: 10.2174/1389203721666200606220109. PMID: 32504500.
- Skelly JV, Madden CB. Overexpression, Isolation, and Crystallization of Proteins. In: Crystallographic Methods and Protocols. New Jersey: Humana Press; 23–54.
- Alias FL, Nezhad NG, Normi YM, Ali MSM, Budiman C, Leow TC. Recent Advances in Overexpression of Functional Recombinant Lipases. Mol Biotechnol. 2023 Nov;65(11):1737-1749. doi: 10.1007/s12033-023-00725-y. Epub 2023 Mar 27. PMID: 36971996.
- Karabchevsky A, Katiyi A, Ang AS, Hazan A. On-chip nanophotonics and future challenges. Nanophotonics. 2020 Sep 11; 9(12):3733–53.
- Zhang N, Ashikuzzaman M, Rivaz H. Clutter suppression in ultrasound: performance evaluation and review of low-rank and sparse matrix decomposition methods. Biomed Eng Online. 2020 May 28;19(1):37. doi: 10.1186/s12938-020-00778-z. PMID: 32466753; PMCID: PMC7254711.
- Qin W, Xie S, Zhang J, Zhao D, He C, Li H. An Analysis on Commercial Screening Kits and Chemical Components in Biomacromolecular Crystallization Screening. Crystal Research and Technology. 2019 Sep 7;54(9).
- Zigon N, Duplan V, Wada N, Fujita M. Crystalline Sponge Method: X-ray Structure Analysis of Small Molecules by Post-Orientation within Porous Crystals-Principle and Proof-of-Concept Studies. Angew Chem Int Ed Engl. 2021 Nov 22;60(48):25204-25222. doi: 10.1002/anie.202106265. Epub 2021 Aug 3. PMID: 34109717.
- Zhou H, Sabino J, Yang Y, Ward MD, Shtukenberg AG, Kahr B. Tailor-Made Additives for Melt-Grown Molecular Crystals: Why or Why Not? Annu Rev Mater Res. 2023 Jul 3;53(1):143–64.
- Liu Z, Zhou Y, Liu J, Chen J, Heck AJR, Wang F. Reductive methylation labeling, from quantitative to structural proteomics. TrAC Trends in Analytical Chemistry. 2019 Sep; 118:771–8.
- Derewenda ZS, Godzik A. The “Sticky Patch” Model of Crystallization and Modification of Proteins for Enhanced Crystallizability. In 2017; 77–115.
- Naderi-Azad S, Croitoru D, Khalili S, Eder L, Piguet V. Research Techniques Made Simple: Experimental Methodology for Imaging Mass Cytometry. Journal of Investigative Dermatology. 2021 Mar; 141(3):467-473.e1.
- Kunachowicz D, Ściskalska M, Jakubek M, Kizek R, Kepinska M. Structural changes in selected human proteins induced by exposure to quantum dots, their biological relevance and possible biomedical applications. NanoImpact. 2022 Apr;26:100405. doi: 10.1016/j.impact.2022.100405. Epub 2022 May 1. PMID: 35560289.
- Barends TRM, Stauch B, Cherezov V, Schlichting I. Serial femtosecond crystallography. Nat Rev Methods Primers. 2022 Aug 4;2:59. doi: 10.1038/s43586-022-00141-7. PMID: 36643971; PMCID: PMC9833121.
- Candoni N, Grossier R, Lagaize M, Veesler S. Advances in the Use of Microfluidics to Study Crystallization Fundamentals. Annu Rev Chem Biomol Eng. 2019 Jun 7;10:59-83. doi: 10.1146/annurev-chembioeng-060718-030312. Epub 2019 Apr 24. PMID: 31018097.
- Deresz KA, Łaski P, Kamiński R, Jarzembska KN. Advances in Diffraction Studies of Light-Induced Transient Species in Molecular Crystals and Selected Complementary Techniques. Crystals (Basel). 2021 Nov 3;11(11):1345.
- Burley SK, Berman HM, Duarte JM, Feng Z, Flatt JW, Hudson BP, Lowe R, Peisach E, Piehl DW, Rose Y, Sali A, Sekharan M, Shao C, Vallat B, Voigt M, Westbrook JD, Young JY, Zardecki C. Protein Data Bank: A Comprehensive Review of 3D Structure Holdings and Worldwide Utilization by Researchers, Educators, and Students. Biomolecules. 2022 Oct 4;12(10):1425. doi: 10.3390/biom12101425. PMID: 36291635; PMCID: PMC9599165.
- Mingos DMP. Early History of X-Ray Crystallography. 2020; 1-41.
- Abdollahi H, Prestegard JH, Valafar H. Computational modeling multiple conformational states of proteins with residual dipolar coupling data. Curr Opin Struct Biol. 2023 Oct;82:102655. doi: 10.1016/j.sbi.2023.102655. Epub 2023 Jul 14. PMID: 37454402.
- Kermani AA. A guide to membrane protein X-ray crystallography. FEBS J. 2021 Oct;288(20):5788-5804. doi: 10.1111/febs.15676. Epub 2020 Dec 31. PMID: 33340246.
- Zheng H, Hou J, Zimmerman MD, Wlodawer A, Minor W. The future of crystallography in drug discovery. Expert Opin Drug Discov. 2014 Feb;9(2):125-37. doi: 10.1517/17460441.2014.872623. Epub 2013 Dec 28. PMID: 24372145; PMCID: PMC4106240.
- Chapuis G. An elementary treatment on the diffraction of crystalline structures. Crystallogr Rev. 2021 Oct 2;27(3–4):146–77.
- Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994 Sep 1;50(Pt 5):760-3. doi: 10.1107/S0907444994003112. PMID: 15299374.
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002 Nov;58(Pt 11):1948-54. doi: 10.1107/s0907444902016657. Epub 2002 Oct 21. PMID: 12393927.
- Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006 Aug;62(Pt 8):859-66. doi: 10.1107/S0907444906019949. Epub 2006 Jul 18. PMID: 16855301.
- Chen B, Zheng W, Chun F, Xu X, Zhao Q, Wang F. Synthesis and hybridization of CuInS2 nanocrystals for emerging applications. Chem Soc Rev. 2023 Nov 10. doi: 10.1039/d3cs00611e. Epub ahead of print. PMID: 37947021.
- Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999 May;6(5):458-63. doi: 10.1038/8263. PMID: 10331874.
- Terwilliger T. SOLVE and RESOLVE: automated structure solution, density modification and model building. J Synchrotron Radiat. 2004 Jan 1;11(Pt 1):49-52. doi: 10.1107/s0909049503023938. Epub 2003 Nov 28. PMID: 14646132.
- Turk D. Towards automatic macromolecular crystal structure determination. Nato Science Series Sub Series I Life and Behavioural Sciences. 2001;325:148–55.
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47 ( Pt 2):110-9. doi: 10.1107/s0108767390010224. PMID: 2025413.
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60(Pt 12 Pt 1):2126-32. doi: 10.1107/S0907444904019158. Epub 2004 Nov 26. PMID: 15572765.
- Chruszcz M, Wlodawer A, Minor W. Determination of protein structures--a series of fortunate events. Biophys J. 2008 Jul;95(1):1-9. doi: 10.1529/biophysj.108.131789. Epub 2008 Apr 25. PMID: 18441029; PMCID: PMC2426657.
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997 May 1;53(Pt 3):240-55. doi: 10.1107/S0907444996012255. PMID: 15299926.
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998 Sep 1;54(Pt 5):905-21. doi: 10.1107/s0907444998003254. PMID: 9757107.
- Patel JR, Joshi H V., A. Shah U, K. Patel J. A Review on Computational Software Tools for Drug Design and Discovery. Indo Global Journal of Pharmaceutical Sciences. 2022; 12:53–81.
- Liebschner D, Afonine PV, Baker ML, Bunkóczi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, Moriarty NW, Oeffner RD, Poon BK, Prisant MG, Read RJ, Richardson JS, Richardson DC, Sammito MD, Sobolev OV, Stockwell DH, Terwilliger TC, Urzhumtsev AG, Videau LL, Williams CJ, Adams PD. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019 Oct 1;75(Pt 10):861-877. doi: 10.1107/S2059798319011471. Epub 2019 Oct 2. PMID: 31588918; PMCID: PMC6778852.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993; 26(2): 283-91.
- Hooft RW, Vriend G, Sander C, Abola EE. Errors in protein structures. Nature. 1996 May 23;381(6580):272. doi: 10.1038/381272a0. PMID: 8692262.
- Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003 Feb 15;50(3):437-50. doi: 10.1002/prot.10286. PMID: 12557186.
- Aguilar FJ, Mills JP, Delgado J, Aguilar MA, Negreiros JG, Pérez JL. Modelling vertical error in LiDAR-derived digital elevation models. ISPRS Journal of Photogrammetry and Remote Sensing. 2010 Jan;65(1):103–10.
- Xie W, Wang F, Li Y, Lai L, Pei J. Advances and Challenges in De Novo Drug Design Using Three-Dimensional Deep Generative Models. J Chem Inf Model. 2022 May 23;62(10):2269-2279. doi: 10.1021/acs.jcim.2c00042. Epub 2022 May 11. PMID: 35544331.
- Wlodawer A, Minor W, Dauter Z, Jaskolski M. Protein crystallography for non-crystallographers, or how to get the best (but not more) from published macromolecular structures. FEBS J. 2008 Jan;275(1):1-21. doi: 10.1111/j.1742-4658.2007.06178.x. Epub 2007 Nov 23. PMID: 18034855; PMCID: PMC4465431.
- Šrajer V, Schmidt M. Watching Proteins Function with Time-resolved X-ray Crystallography. J Phys D Appl Phys. 2017 Sep 20;50(37):373001. doi: 10.1088/1361-6463/aa7d32. Epub 2017 Aug 22. PMID: 29353938; PMCID: PMC5771432.
- Wlodawer A, Sekula B, Gustchina A, Rotanova TV. Structure and the Mode of Activity of Lon Proteases from Diverse Organisms. J Mol Biol. 2022 Apr 15;434(7):167504. doi: 10.1016/j.jmb.2022.167504. Epub 2022 Feb 17. PMID: 35183556; PMCID: PMC9013511.
- Rotanova TV, Botos I, Melnikov EE, Rasulova F, Gustchina A, Maurizi MR, Wlodawer A. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 2006 Aug;15(8):1815-28. doi: 10.1110/ps.052069306. PMID: 16877706; PMCID: PMC2242575.
- Wlodawer A, Sekula B, Gustchina A, Rotanova TV. Structure and the Mode of Activity of Lon Proteases from Diverse Organisms. J Mol Biol. 2022 Apr 15;434(7):167504. doi: 10.1016/j.jmb.2022.167504. Epub 2022 Feb 17. PMID: 35183556; PMCID: PMC9013511.
- Rotanova TV, Botos I, Melnikov EE, Rasulova F, Gustchina A, Maurizi MR, Wlodawer A. Slicing a protease: structural features of the ATP-dependent Lon proteases gleaned from investigations of isolated domains. Protein Sci. 2006 Aug;15(8):1815-28. doi: 10.1110/ps.052069306. PMID: 16877706; PMCID: PMC2242575.