More Information
Submitted: May 12, 2021 | Approved: May 25, 2021 | Published: May 26, 2021
How to cite this article: Saibi W, Brini F. Ion transporters and their molecular regulation mechanism in plants. J Plant Sci Phytopathol. 2021; 5: 028-043.
DOI: 10.29328/journal.jpsp.1001058
Copyright License: © 2021 Saibi W, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Ion Transporters; Na+ sensing; Na+ transport; Potassium; Proton Pumps; Salinity
Ion transporters and their molecular regulation mechanism in plants
Walid Saibi and Faiçal Brini*
Laboratory of Biotechnology and Plant Improvement, Centre of Biotechnology of Sfax, BP1177, 3018 Sfax, Tunisia
*Address for Correspondence: Faiçal Brini, Laboratory of Biotechnology and Plant Improvement, Centre of Biotechnology of Sfax, BP1177, 3018 Sfax, Tunisia, Email: [email protected]
With the global population predicted to grow by at least 25% by 2050, the need for sustainable production of nutritious foods is important for human and environmental health. Recent progress demonstrate that membrane transporters can be used to improve yields of staple crops, increase nutrient content and resistance to key stresses, including salinity, which in turn could expand available arable land. Exposure to salt stress affects plant water relations and creates ionic stress in the form of the cellular accumulation of Na+ and Cl− ions. However, salt stress also impacts heavily on the homeostasis of other ions such as Ca2+, K+, and NO3- and therefore requires insights into how transport and compartmentation of these nutrients are altered during salinity stress. Since Na+ interferes with K+ homeostasis, maintaining a balanced cytosolic Na+/K+ ratio has become a key salinity tolerance mechanism. Achieving this homeostatic balance requires the activity of Na+ and K+ transporters and/or channels. The aim of this review is to seek answers to this question by examining the role of major ions transporters and channels in ions uptake, translocation and intracellular homeostasis in plants.
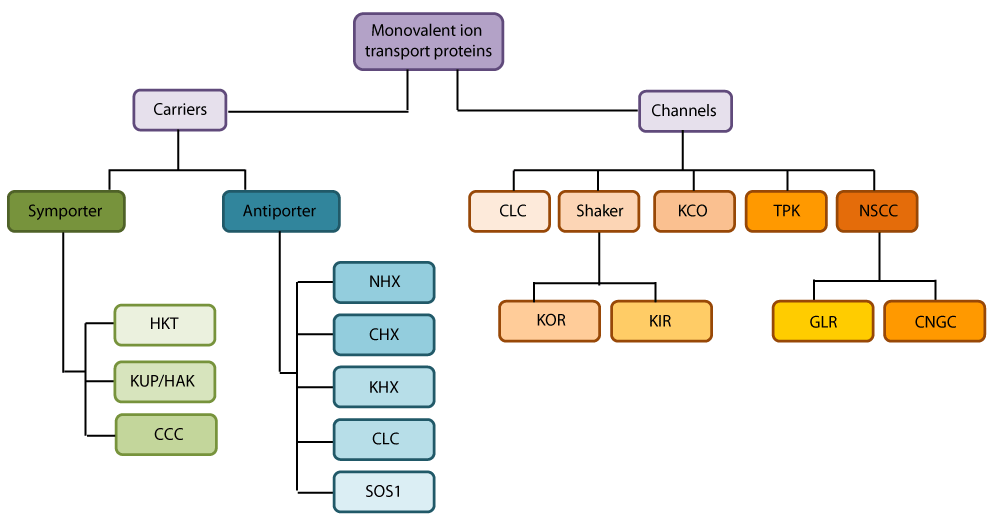
The beginning of 21st century is marked by global scarcity of water resources, environmental pollution and increased salinization of soil and water [1]. Increasing human population and reduction in land available for cultivation are two threats for agricultural sustainability [2]. A saline soil is generally defined as one in which the electrical conductivity (EC) of the saturation extract (ECe) in the root zone exceeds 4 dS m−1 (approximately 40 mM NaCl) at 25 °C and has an exchangeable sodium of 15%. The yield of most crop plants is reduced at this ECe, though many crops exhibit yield reduction at lower ECes [3,4]. It has been estimated that worldwide 20% of total cultivated and 33% of irrigated agricultural lands are afflicted by high salinity. Furthermore, the salinized areas are increasing at a rate of 10% annually for various reasons, including low precipitation, high surface evaporation, weathering of native rocks, irrigation with saline water, and poor cultural practices. It has been estimated that more than 50% of the arable land would be salinized by the 2050 [4]. To meet the projected food demand of 9.3 billion people by 2050, global agricultural production must be increased by 60 % from its 2005–2007 levels [5]. This urgent need requires a large effort to improve agricultural production. One feasible way to cope with this challenge is to breed robustly salt-tolerant crops. Understanding the mechanisms underlying plant salt tolerance would be of benefit for breeding such crops and mitigating future food shortages. Accumulation of high Na+ in the cytosol can not only cause K+ deficiency and thus disrupt various enzymatic processes, but also impose an energetic burden on the cell owing to the requirement of organic solute synthesis to compensate for the export of Na+ for osmotic adjustment [6]. Thus, understanding how Na+ is sensed and transported in plants under saline conditions could help researchers or breeders breed crops with robust salt tolerance. The present review is focused on the main processes that contribute to the overall homeostasis of the main ionic constituents of salinity and also analyses which specific membrane transporters are believed to be involved in uptake, extrusion, long distance transport and compartmentalization of salt at the cellular and tissue level. Figure 1 gives an overview of the main classes of monovalent ion transporters that totals hundreds of isoforms, often derived from large gene families. In the following sections, we will analyze the potential roles of transporter classes and specific proteins regarding uptake, efflux translocation and compartmentation of salt. In addition, these sections will also evaluate which of these provide promising targets in the quest to improve crop salt tolerance.
Na+ sensing in plants
Possible salt sensors for perception of Na+: Unlike in animal cells, no specific salt sensors have been identified in plant cells to date. Thus, our knowledge of how plants perceive salt stress and thus decode the corresponding signals remains limited. Cramer, et al. [7] found that Ca2+ can mitigate the loss of membrane integrity and minimize cytosolic K+ leakage and proposed that displacement of Ca2+ by Na+ from the root cell plasmalemma is a primary response to salt stress. However, Kinraide [8] showed that the Ca2+-displacement hypothesis is often of minor importance to salt stress response. SOS1 (salt overly sensitive 1) Na+/H+ antiporters [9], histidine kinases [10], and AHK1/ATHK1 [11] have also been suggested to be potential salt sensors or osmo-sensors. Shabala, et al. [12] suggested some putative salt stress sensors/proteins involved in early signaling events, including exchangers and transporters such as SOS1 Na+/H+ antiporters, NCX Na+/Ca2+ exchangers, NSCC/NADPH oxidase tandem, mechanosensory channels and transporters, cyclic nucleotide receptors, purino-receptors, annexins, and H+-ATPase/GORK tandem. The binding of salt stress-induced increases of cyclic nucleotides to their receptors, e.g. CNGCs, can activate this CNGC Ca2+-permeable channels, and thus the increase of cyclic nucleotides could be translated into a massive cytosolic Ca2+ uptake, which can affect Ca2+ signaling [12]. Similarly, sensing of salt-induced eATP (extracellular ATP) by plasma membrane purinoceptors can be translated into other signaling events, such as ROS (reactive oxygen species) and cytosolic Ca2+ signature [13].
Figure 1: Overview of main gene families involved in Na+, K+ and Cl− homeostasis in crops plants during salt stress. Abbreviations: CCC, cation chloride co-transporter; CHX, cation/H+ exchanger; CLC, voltage gated Cl− channel; CNGC, cyclic nucleotide gated channel; GLR, glutamate like receptor; HKT, high affinity K+ transporter; KCO, K+ outward rectifying channel; KHX, K+/H+ exchanger; KIR, Shaker type K+ inward rectifier; KOR, Shaker type K+ outward rectifier; KUP/HAK, K+ uptake permease; NHX, Na+/H+ exchanger; NSCC, non-selective cation channel; TPK, two-pore K+ channel.
Root meristem zone: a tissue harboring salt sensors?: Root is the first plant organ that encounters salinity. Thus, Na+ enters first into roots and is then transported to shoots. Wu, et al. [14] found that salt-tolerant bread wheat varieties had significantly higher cytosolic Na+ in the root meristem zone than salt-sensitive varieties; although no difference in vacuolar Na+ fluorescence intensity was found in the root meristem zone. This finding suggests that salt-tolerant wheats could have more ability to buffer or tolerate increased Na+ in the cell cytosol in root meristem zone than salt-sensitive wheats. Further, by removal of the root meristem zone from salt-tolerant wheat varieties, Na+ distribution in mesophyll cells was altered and a salt-sensitive phenotype resulted [15]. Taken together, these findings suggest that the root meristem zone can act as a salt stress sensor, or at least a tissue that harbors salt stress-sensor components.
Transporters and channels involved in na+ transport in plants under salt stress
The importance of Na+ exclusion in plant salt tolerance: The importance of Na+ exclusion in protecting plants against salinity stress is widely accepted. Under salt stress, net Na+ accumulation in plant cells is determined by the ion-exchange activity of Na+ influx and efflux. Na+ influx occurs mainly through ion channels such as the high-affinity K+ transporter HKT and non-selective cation channels (NSCC), and Na+ efflux is known to be mediated by SOS1, a Na+/H+ antiporter. In the presence of elevated levels of external Na+, under saline conditions, Na+ efflux from plant cells is an active process [16]. To date, SOS1, expressed mainly in the root apex in Arabidopsis [17], is the only transporter that has been characterized in Na+ export from the cytosol to the apoplast. Loss of SOS1 function resulted in a hyper-salt-sensitive phenotype in the halophytic Arabidopsis relative Thellungiella salsuginea [18]. This finding further confirmed the important role of the SOS1 Na+/H+ antiporter in Na+ exclusion and overall plant salt tolerance. Moreover, to date, studies showing the important role of Na+ exclusion in overall salt tolerance have been based mostly on shoot/leaf or even whole-plant Na+ content [19-24]. Whether this restricted Na+ accumulation in shoot/leaves is achieved mainly by root Na+ export or shoots Na+ exclusion, or by both of these processes with tight regulation/coordination at different growth stages and time scales, however, has remained unclarified.
The importance of vacuolar Na+ sequestration in plant salt tolerance: SOS1-mediated Na+ export from cytosol to apoplast (against Na+ gradient) is an energy-consuming process. Given that most of the cell volume is occupied by vacuole and most metabolisms occurs in the cytoplasm, one way for plants to alleviate Na+ toxicity in the cytosol is to store Na+ in the vacuole. Vacuolar Na+ sequestration is a common and important mechanism in plant salt tolerance, and is mediated by Na+/H+ antiporters [25-27]. Prevention of cytoplasmic Na+ elevation, maintenance of the cytosolic K+/Na+ ratio, and control of vacuolar osmotic potential in plants under salt stress can be achieved by, or is associated with, vacuolar Na+ sequestration [28]. To date, the best-known transporter for vacuolar Na+ sequestration is the NHX1 Na+, K+/H+ exchanger. Overexpression of NHX1 improves salt tolerance in many species including Arabidopsis [25], tomato [29], rice [30], and tobacco [31], showing the importance of vacuolar Na+ sequestration in plant overall salt tolerance. Salt-tolerant wheat varieties showed significantly higher vacuolar Na+ fluorescence intensity in mature root cells than did sensitive varieties [14,32]. Under overexpression of OsNHX1, transgenic rice cells survived better under saline condition and showed significantly higher growth rate and total Na+ content than the wild type (WT) [33]. Taken together, these findings show clearly that vacuolar Na+ sequestration is an important trait contributing to plant overall salt tolerance. After sequestration of Na+ in vacuoles, another important concern is to prevent Na+ leakage from vacuole to cytosol. Loss of control of this step could result in futile Na+ cycling between vacuole and cytosol, imposing a high energy burden on the plant. FV (fast-activating) and SV (slow-activating) channels are tonoplast Na+ and K+-permeable channels that control Na+ leakage from vacuole to cytosol. Negative control of FV and SV channel activity has been shown in the salt-stressed halophyte quinoa to reduce such leakage [34], suggesting that efficient control of Na+ leakage from vacuole to cytosol could be an important mechanism in plant overall salt stress tolerance (Figurer 2).
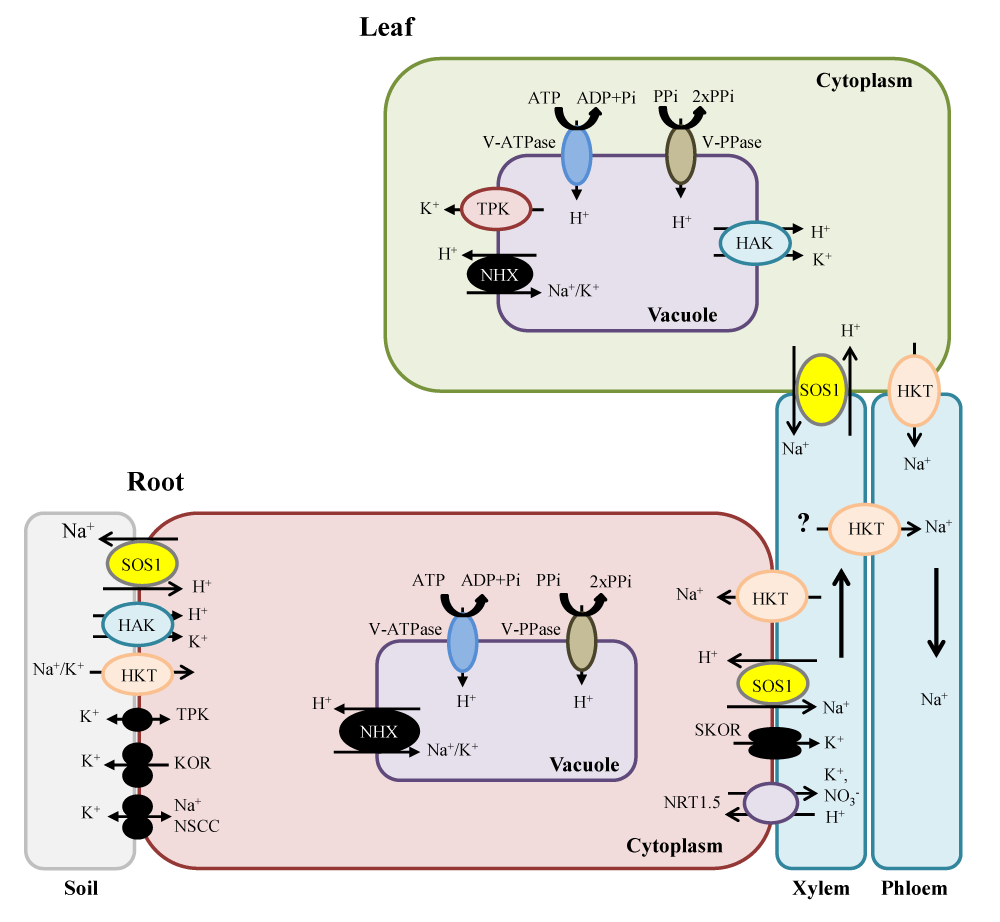
Figure 2: Schematic representation showing key plasma and tonoplast membrane transporters, channels and pumps mediating Na+ and K+ homeostasis in plants under salt stress. Na+ ions enter the cells via Non-Selective Cation Channels (NSCCs) and possibly via other cation transporters (symplast flow) and through the cell wall and intercellular spaces (apoplast flow). The Na+/H+ antiporter SOS1 extrudes Na+ at the root soil interface, thus reducing the Na+ net influx of Na+. At the xylem parenchyma cells, HKT1-like proteins retrieve Na+ from the xylem sap, thereby restricting the amount of Na+ reaching the photosynthetic tissues. To translocate Na+ back to the root, ions unloaded from xylem may be transported into phloem via additional HKT1-like protein. In addition, HKT1-like proteins also load Na+ into shoot phloem, and then Na+ is transferred into roots via phloem, preventing Na+ accumulation in shoots. SOS1, localized in the xylem parenchyma cells, is also suggested to mediate Na+ efflux from xylem vessels under high salinity. Incoming Na+, in root and shoots, is stored in the large central vacuole by tonoplast-localized NHX exchangers. Plasma membrane (PM) H+-ATPase (P-ATPase), PM H+-PPase (PM-PPase), tonoplast H+-ATPase (V-ATPase) and tonoplast H+-PPase (V-PPase) generate electrochemical potential gradient for secondary active transport.
Control of xylem Na+ loading and unloading: Roots absorb ions and then transfer them to shoots via xylem loading, so that control of xylem Na+ loading is important in plant overall salt tolerance. To date, SOS1 Na+/H+ antiporter [16,35,36], CCC co-transporter [37], and SKOR channel (if xylem loading of Na+ is passive) [38] have been shown to be involved in xylem Na+ loading (Figure 2). Shi, et al. [16] suggested that SOS1 plays a role in xylem Na+ loading in Arabidopsis under mild salt stress. Yadav, et al. [39] showed that enhanced xylem Na+ loading and higher overall salt tolerance was achieved in tobacco by overexpression of SbSOS1. Recently, a reduction in overall net xylem Na+ loading and accumulation in the shoot and thus improved salt tolerance were observed in wheat Nax (locus for Na+ exclusion) lines following down regulation of SOS1-like Na+/H+ antiporter [40]. Besides SOS1, a CCC co-transporter that is preferentially expressed at the xylem/symplast boundary has also been suggested to play a role in xylem Na+ loading [37] (Figure 2). With respect to Na+ transport in xylem, besides Na+ loading into xylem, Na+ unloading from xylem is another important mechanism. HKT transporters play a main role in this process. Sunarpi, et al. [41] showed that the AtHKT1 transporter located on the plasma membrane in xylem parenchyma cells in leaves played a role in Na+ unloading from xylem vessels to parenchyma cells. Huang, et al. [42] suggested that TmHKT7-A2, which is associated with Nax1 locus, could control xylem Na+ unloading in roots and sheaths. Also, Byrt, et al. [20] showed that HKT1;5 is strongly associated with Nax2 locus in durum wheat and its orthologous locus Kna1 in bread wheat removes Na+ from xylem in roots and leads to a high K+/Na+ ratio in leaves. Jaime-Perez, et al. [43] showed that the SlHKT1;2 Na+-selective transporter plays an important role in Na+ unloading from xylem in tomato shoots and thus modulates its Na+ homeostasis under salinity (Figure 2).
Na+ recirculation from shoot to root via phloem: Na+ recirculation from shoots to roots has been suggested as an efficient way to protect leaf cells from Na+ toxicity [44]. Because leaf vacuolar Na+ sequestration ability is poor, Na+ recirculation from shoots to roots via phloem sap is probably the main mechanism involved in prevention of Na+ delivery to leaf cells in most salt-sensitive plants [45]. Apart from shoot growth rate, the rate of recirculation of Na+ to the roots via phloem has been suggested as an important factor affecting Na+ concentrations in shoots [46]. In several species, such as lupine, clover, sweet pepper, and maize, recirculation of Na+ to roots via phloem played a role in overall salt tolerance [47]. Berthomieu, et al. [48] showed that expression of the AtHKT1 gene was restricted to phloem tissues in all organs in Arabidopsis, and that the AtHKT1 gene was involved in Na+ recirculation from shoots to roots probably by mediating Na+ loading into phloem sap in the shoots and unloading it in roots. However, in Arabidopsis, a role of AtHKT1 in control of both Na+ accumulation in roots and retrieval of Na+ from xylem, without involvement in root influx or recirculation in the phloem, was suggested by Davenport, et al. [46]. Ren, et al. [49] showed that HKT-type transporter encoded by SKC1 (shoot K+ concentration 1) gene might be involved in the recirculation of Na+ by unloading it from the xylem in rice. Kobayashi, et al. [50] found that an OsHKT1;5 Na+ selective transporter associated with the SKC1 locus is localized in cells adjacent to the xylem in roots, and is involved in mediating Na+ exclusion in phloem to protect young leaf blades of rice under salt stress (Figure 2).
Na+ transporters: To date, most members of the cation/proton antiporter (CPA) family have been identified as Na+/H+antiporters (subclass 1), but a few are K+/H+ antiporters, including CHX13, CHX17, CHX20, and CHX23 in the CPA2 family [51]. Besides vacuolar Na+ sequestration, another important pathway for controlling Na+ distribution in plant cells is Na+ exclusion/export. To date, SOS1 Na+/H+ antiporter is the only reported antiporter responsible for Na+ export from plant cells [52,53]. SOS1 activity is regulated by SOS2, a serine/threonine protein kinase (CIPK24) and SOS3, a myristoylated calcium-binding protein (CBL4) [54-56]. SOS3 recruits SOS2 to the plasma membrane, and then this CBL-CIPK complex activates SOS1 by phosphorylation, dramatically increasing Na+/H+ exchange activity (Figure 3) [16]. Moreover, the existence of an ATP-driven Na+ transport mediated by a Na+-ATPase at the plasma membrane has been shown in lower plants, such as the marine alga Heterosigma akashiwo [57] and the moss Physcomitrella patens [58].
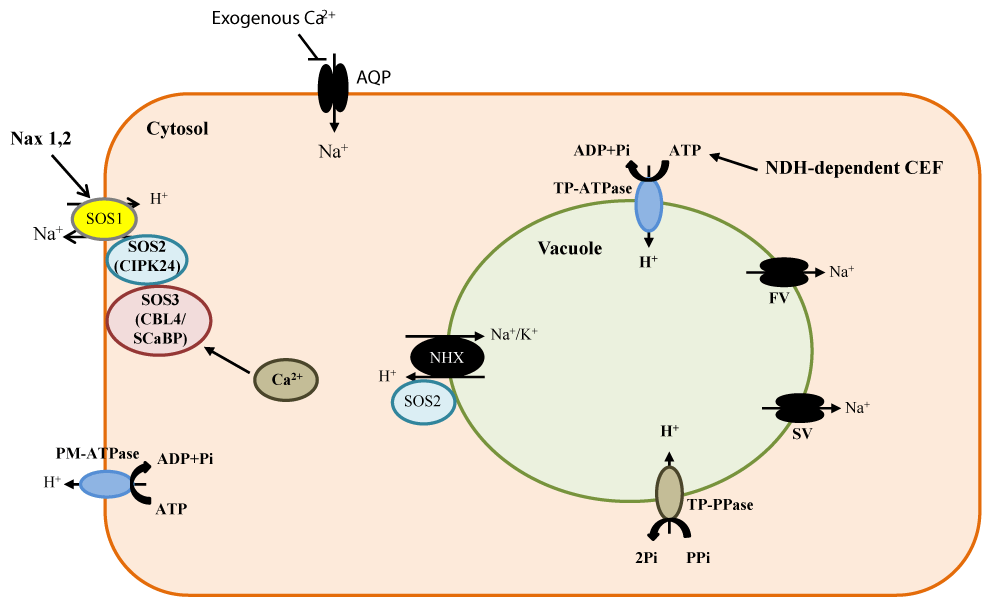
Figure 3: Intracellular Na+ homeostasis mediated by Na+ transporters and channels and their regulatory elements. Some new components in these transport mechanisms have been added, i.e., the regulation of stelar-localized SOS1 activity by Nax1 and Nax2 Na+ exclusion loci in rice. This regulation improves salt stress tolerance by enhancing the retrieval of Na+ from xylem back into stellar cells. Another component is the potential involvement of plant aquaporins (AQP, AtPIP2;1 in particular) in Na+ uptake. Equally important is the role of FV (fast vacuolar) and SV (slow vacuolar) channels that mediate vacuolar Na+ leakage to the cytosol, deemed a salt-sensitive trait. Worthy of note also is the role of PM and TP H+ pumps that generate a pmf to energize Na+ transport via the two Na+/H+ exchangers (SOS1 and NHXs), The NADPH dehydrogenase (NDH)-dependent cyclic electron flow (CEF) constitutes an important source of ATP required to fuel Na+ sequestration into vacuoles.
The role of HKT1: X transporters in Na+ unloading and recirculation in salt stressed plants was mentioned in the previous sections. For example, Kobayashi, et al. [50] found that the OsHKT1;5 Na+ selective transporter, which is associated with the SKC1 locus, is localized in cells adjacent to the xylem in roots, and is involved in mediating Na+ exclusion in phloem to protect young leaf blades of rice under salt stress.
Na+ channels: NSCCs are a large family of channels that lack selectivity for cations. They are typically permeable to wide range of monovalent cations [59] and are located on both the plasma membrane and the tonoplast (Figure 2). They can be divided into voltage-dependent NSCCs (depolarization-activated, hyper-polarization-activated), voltage-independent NSCCs, ROS-activated NSCCs, amino acid-activated NSCCs, cyclic nucleotide-gated NSCCs, etc. Electrophysiological studies suggest that Na+ influx across the plasma membrane occurs via NSCC/VIC in root cortical cells [16,60,61]. Maathuis and Sanders [62] found that cyclic nucleotide-regulated VIC (voltage-independent cation channels) channels showed no selectivity among monovalent cations in Arabidopsis root cells.
Molecular regulation of Na+ trans Moreover, the existence of an porters/channels in response to salt stress: To date, SOS1 is the only known anti-transporter responsible for Na+ export from cytosol to apoplast. Usually, expression of the SOS1 gene is up-regulated in salt stressed plants [52,63,64]. The functional activity of SOS1 mediated Na+ export could be influenced by SOS2 [54], SOS3 [56], the assembly of SOS2-SOS3 complex [65], and H+-ATPase, which can increase H+ efflux to energize Na+ efflux through SOS1 antiporters [66]. SOS1 activity could also be influenced by ROS or ROS signaling-associated components. SOS1 mRNA stability is increased in Arabidopsis under H2O2 treatment, and NADPH oxidase is also involved in the up-regulation of SOS1 mRNA stability [67]. Besides, SOS1 interacts with RCD1 (radical-induced cell death), a regulator of oxidative stress responses, and functions in oxidative stress tolerance in Arabidopsis [68]. Reduced ROS production and increased SOS1 expression was found in pao1pao5 (polyamine oxidase, PAO) Arabidopsis mutants than in the WT under salt stress [69]. As with SOS1, overexpression of NHX1 to increase plant salt tolerance has been shown in many plant species [25,70,71]. Although the role of AtNHX1 in K+ accumulation in the vacuole was discovered in recent years [72-74], this finding cannot completely rule out the involvement of NHX1 in vacuolar Na+ sequestration, especially under high salinity [36,75]. Usually, the NHX1 gene is up-regulated in salt-stressed plants, including Arabidopsis [76], barley [77], and alfalfa [78]. However, a clear decrease in the transcript level of NHX1 in wheat roots was observed under salt stress, while almost no change in the NHX1 transcript level was found in leaves [79]. Moroever, in contrast to the successfully improved salt stress tolerance in tomato [29], rice [30], and tobacco [31], overall salt tolerance was not enhanced in Arabidopsis [74] and barley [78] by expression of the NHX1 Na+/H+ exchanger gene. These conflicting results raise the questions of the importance of tissue specificity in plant salt-stress tolerance. NHX1 is known to be fueled by an H+ gradient across the tonoplast that is maintained by vacuolar H+-ATPase and vacuolar PPase [80]. Expressing a halophyte vacuolar H+-ATPase subunit c1 (SaVHAc1) in rice plants resulted in higher chlorophyll content and yield than in its WT [81]. Overexpression of vacuolar PPase AVP1 improved salt tolerance in transgenic Arabidopsis relative to the WT, showing a healthy growth of transgenic Arabidopsis in the presence of 250 mmol L−1 NaCl compared with the WT, which died after 10 days [82]. These results suggest that manipulating vacuolar H+-ATPase and PPase could allow regulating NHX1 activity and eventually plant overall salt tolerance. Other known factors in the regulation of NHX1 activity are SOS2 [83] and CaM15 [84]. Also, CBL10 can interact with SOS2 to protect Arabidopsis shoots from salt stress [85]. Tang, et al. [86] showed that PtCBL10A and PtCBL10B interact with PtSOS2 in the vacuolar membrane to regulate shoot salt tolerance in poplar. Thus, CBL10 is also proposed to regulate NHX1 activity [87]. Two recent reviews have also focused on molecular regulation of Na+ transporters/channels in response to salt stress [36,88].
Transporters involved in cl- uptake
Cl− is a major solute in plant vacuoles, particularly during salt stress, and is involved in both turgor and osmoregulation [89]. Although there is a substantial amount of information regarding K+ and Na+ transport in plants, very little is known about the molecular mechanism behind the substantial Cl− influx that results from salinization [90]. Plants contain CLC type anion channels which are believed to participate in turgor regulation, stomatal movement and anionic nutrient transport such as NO3- [91]. Although the transcript abundance of several CLCs is affected by salinity [92], they are unlikely to contribute to root Cl− uptake: Firstly, plant CLCs have only been detected at endomembranes which appears to exclude a role in Cl− uptake and secondly the thermodynamics of Cl− uptake rule out passive channel type mechanisms. A second class of potential Cl− transporters is formed by the cation chloride cotransporters (CCCs) encoding one gene in Arabidopsis and two genes in rice. AtCCC, expressed in root and shoot tissues, probably functions as a 2Cl−:K+:Na+ cotransporter. Loss of function of AtCCC in Arabidopsis led to a changed root: shoot Cl− ratio but also to a large increase in net Cl− uptake arguing against a role of AtCCC in the uptake of this ion [37].
In addition to Na+, Cl− compartmentation is also important for salt tolerance, as elevated levels of Cl− in the cytosol may be harmful, particularly in the case of citrus [93]. Since the vacuole is moderately positive with reference to the cytoplasm, part of the vacuolar Cl− sequestration could proceed through ion channels and several voltage-gated anion channels of the CLC family have been detected in the tonoplast of various species. In Arabidopsis, CLCa was recently shown to function primarily as a H+ coupled antiporter to drive vacuolar nitrate accumulation [94], whereas CLCc may also be involved in NO3- homeostasis rather than vacuolar Cl− sequestration. However, CLC transcription has been found to respond to salinity: In rice, OsCLCa was significantly upregulated in salt sensitive cultivars in response to salinity stress and OsCLCc, which is expressed in both leaves and roots, showed transcript reduction in the chloride accumulating salt tolerant Pokkali variety [95]. Diédhiou and Golldack [92] showed a coordinated regulation of anion and cation homeostasis in salt-treated rice and suggested a function for OsCLCc in osmotic adjustment at high salinity. A similar co-regulation was recorded in soybean for NHX1 and CLC1 [96]. Nakamura, et al. [97] showed that the same CLC channels partially complemented the yeast gef1 mutant which lacks the yeast CLC channel. In conclusion, these findings suggest that CLC type anion channels are important in mediating Cl− sequestration in the vacuole (Figure 2).
Potassium transporters in plants: involvement in k+ acquisition, redistribution and homeostasis
Potassium is a major plant nutrient which has to be accumulated in great quantity by roots and distributed throughout the plant and within plant cells. Membrane transport of potassium can be mediated by potassium channels and secondary potassium transporters. Uptake and distribution of K+ in plant cells is carried out by a variety of transporter proteins categorized into several families with varied structures and transport mechanisms that comprise the channel families Shaker-like voltage-dependent, the tandem-pore (TPK), and the two-pore channels (TPC) [98], the carrier-like families KT/HAK/KUP [99,100], HKT uniporters and symporters [101], and cation-proton antiporters (CPA). The CPA family is the largest one and includes the NHX, CHX, and KEA antiporters [102] (Figure 2).
K+-Selective Channels: The first K+ transporter with a role in nutrient uptake was the Shaker-like, voltage-gated, and K+-selective channel AKT1 [103]. Plant voltage-gated K+ channels are divided into three subfamilies regarding their response to the membrane potential [104]: (1) Inward-rectifying (Kin) channels that in Arabidopsis include AKT1, AKT6, KAT1, and KAT2; they open at hyperpolarized membrane potentials allowing the uptake of K+. (2) Outward-rectifying (Kout) channels that mediate K+ release because they open at depolarized membrane potentials; this group is composed of SKOR and GORK channels. (3) Weakly rectifying (Kweak) channels that can mediate both K+ uptake and release, and whose Arabidopsis representative is AKT2. In addition, the Arabidopsis KC1 (KAT3) is an electrically silent Shaker-like protein that interacts with and regulates functionality of the Kin channels AKT1, KAT1, KAT2, and AKT2, but not the Kout channels [105].
K+ non-Selective Channels: Electrophysiological recordings of channel activities in the tonoplast have identified fast vacuolar (FV), slow vacuolar (SV), and K+-selective vacuolar (VK) cation channels that mediate the release of vacuolar K+ [98]. The VK currents have been assigned to two-pore K+ (TPK) channels [106]. TPK1, 2, 3, and 5 of Arabidopsis are located in the tonoplast, while TPK4 is in plasma membrane. TPK1 currents are independent of the membrane voltage but sensitive to cytosolic Ca2+ and regulated by calcium-dependent protein kinases (CDPKs) and 14-3-3 protein binding (Figure 3) [107]. In Arabidopsis, the TPC1 channel accounts for the SV current [108]. TPC1 is voltage-dependent and non-selective, allowing K+ and Na+ to permeate toward the cytosol. Whether TPC1 also permeates Ca2+ or Ca2+ is only an effector of TPC1 gating is a matter of controversy [109]. TPC channels are activated by a decrease in transmembrane potential and increased cytosolic Ca2+, and inhibited by low luminal pH and Ca2+. The ubiquitous nature of TPC channels and the magnitude of the SV/TPC currents are such that TPC channels are capable of contributing substantially to cellular K+ homeostasis. However, plants lacking TPC1 function are not impaired in growth and development. This may indicate that the TPC1 channel is closed most of the time and opens upon specific inputs or under stress. Current thinking is that TPC1 is part of a Ca2+/ROS relay that propagates stress signals [110,111].
KT/HAK/KUP transporters: Proteins of the KT/HAK/KUP family are present in plants, fungi, bacteria, and even viruses [112,113], and they are often associated with K+ transport across membranes and K+ supply. Members of this family have been widely associated with high-affinity K+ uptake from the soil, while others may function in both low-affinity and/or high-affinity transport [59,114] and other roles related, for example, to K+ translocation, control of water mouvement at the plant level, salt tolerance, osmotic/ drought responses, transport of other alkali cations, and developmental processes in plants, such as root hair growth and auxin distribution [100,113]. These diverse functions of KT/HAK/KUP transporters may all result from their critical roles in cellular K+ homeostasis (Figure 2).
HKT uniporters and symporters: The High affinity K+ Transporters (HKTs) facilitate Na+-selective uniport or Na+-K+ symport with a channel-like activity [115] (Figure 2).
Phylogenetic and functional analyses distinguished two HKT subfamilies [116]. Members of subfamily 1 (HKT1) are ubiquitous in plants, Na+-selective, and mostly involved in Na+ recirculation through vascular tissues, as best exemplified by AtHKT1;1 [41]. Members of subfamily 2 (HKT2) have been found only in monocotyledonous species. Although they are all K+-permeable, mechanistically HKT2s can operate as either Na+-K+ symporters or K+-selective uniporters (reviewed by Benito, et al. [115]). HKT2-like proteins of cereals have been involved in K+ nutrition.
Cation-proton antiporters (CPA): The recent meta-analysis of a large number of publications reporting tolerance phenotypes imparted by exchangers of the Cation/Proton Antiporter Family 1 (CPA1, which includes NHX proteins) concluded that the effect on K+ status was generally more pronounced than on Na+ content [117]. An informative work showed that overexpression of the AtNHX1 in tomato induced K+-deficiency symptoms despite transgenic plants having greater K+ contents than controls [72]. The intense sequestration of K+ in NHX1-overexpressing plants reduced cytosolic K+ activity, primed the induction of the high-affinity K+ uptake system, and elicited an array of metabolic and hormonal disorders related to K+ deprivation [72,118]. Notwithstanding these unintended effects resulting from NHX overexpression, NHX proteins do increase salt tolerance, presumably because retention of cellular K+ is a requisite for adaptation to a saline environment [119]. Deletion of NHX1 and NHX2 genes encoding the two majors vacuolar NHX isoforms resulted in the inability to compartmentalize K+ and, surprisingly, in sensitivity to K+ supply at concentrations that did not compromise the growth of control plants [73,74]. Moreover, nhx1 nhx2 mutant lines showed dysfunctional stomatal activity, with impaired opening and closure [74,120].
Long-distance transport and inter-organ K+ partitioning: Potassium absorbed by peripheral root cells and not compartmentalized in vacuoles must be transported to the upper parts of the plant through the xylem [121]. This step is critical in the long-distance distribution of K+ from roots to the upper parts of the plant, and is driven by negative pressure (pulling) created by evaporation of water from leaves. The osmotic water uptake that is caused by nutrient absorption in the root also provides a positive force, known as root pressure, from roots to xylem vessels. Under regular K+ supply, symplastic K+ diffusion to the xylem through the stele may contribute sufficiently to K+ transport from root to shoot [122]. Moreover, K+ is highly mobile within plants, exhibiting cycling between roots and shoots via xylem and phloem [121]. Potassium channels SKOR and AKT2 play an important role in K+ translocation via xylem and phloem. SKOR (Stelar K+ Outward Rectifier), being an outward-rectifying channel, is expressed in root stele cells (pericycle and xylem parenchyma cells) of Arabidopsis, where it mediates K+ secretion by xylem parenchyma cells of roots and toward the xylem vessels [123] (Figure 2). SKOR opens upon membrane depolarization to allow cytosolic K+ efflux. In the presence of ample external K+, the channel opens at less negative membrane voltages, thereby minimizing the risk to serve as an undesirable K+- influx pathway [124]. Upon acute depolarization of plasma membrane induced by salinity, SKOR in xylem parenchyma cells can be rapidly activated to mediate K+ loading into the xylem. After the plasma membrane potential is restored by increased H+-ATPase activity, SKOR-dependent K+ release from root stelar cells to the xylem by membrane depolarization is suppressed. Then, accumulated ROS under salinity could, in turn, activate SKOR channels to allow xylem K+ loading. This may require a highly coordinated mechanism to ensure efficient xylem K+ loading in salt-stressed plants (Figure 2).
Large quantities of K+ recirculate from roots to shoots via the xylem and subsequently return to the roots via the phloem [125,126]. The magnitude of the K+ flux recirculated from the shoots to the roots would constitute a signal by which the growing shoots could communicate to roots for their K+ requirement and regulate K+ secretion into the xylem sap (and eventually root K+ uptake). AKT2 is mainly expressed in the phloem both in leaves and roots [127,128], where the AKT2 channel protein plays a dual role by loading K+ in source tissues and unloading K+ in sink organs [129]. AKT2 is the only weak inward-rectifier characterized in Arabidopsis [130,131]. The protein phosphatase PP2CA interacts with AKT2 to induce both inhibition of the channel current and enhancement of its inward rectification [132]. AKT2 can modulate the membrane voltage by switching between its modes of an inward or a non-rectifying channel, respectively [127,133]. Depending on the cellular context, the phosphorylation status of the AKT2 channels may change, enabling them to drive either inward or outward K+ fluxes [134].
Members of the KT/HAK/KUP family, e.g. AtKUP7 and OsHAK5, have been proposed to facilitate long-distance K+ transport from root to shoot, presumably by mediating K+ uptake into the xylem parenchyma cells [122,135] (Figure 2). This function of KT/HAK/KUP transporters would be relevant under K+ deprivation, when apoplastic K+ levels could be below the operational range of channels.
As mentioned earlier, HKT channel-like proteins are primarily involved in Na+ fluxes both in roots (monocots) and vascular bundles (monocots and dicots) [101]. However, they often have a significant impact on maintaining high K+/Na+ ratio in aerial parts during salinity stress and genetic diversity in HKT proteins meditating long-distance transport of Na+ and K+ have a great impact on the salt tolerance of cereals (Figure 2) [23,49,136].
Co-regulation of k+ and nitrogen uptake
Plants take up numerous mineral nutrients from the soil; some of them are essential (as K+ or NO3−), while others can be toxic at high concentrations (as Na+ or NH4+). Adaptive responses to varying mineral nutrient conditions in the soil, particularly low-nutrient environments, involve multiple signaling pathways whose integration allows plants to grow and adjust their development to each specific nutritional situation [137]. Thus, changes in the concentration of one nutrient trigger a signaling cascade that modify not only the amount, localization, and/or activity of this nutrient-specific transporter/channel, but also transporters/ channels related with other nutrients. N-K interactions are important for root architecture [137].
K+ is the preferred counter ion for root-to-shoot translocation of NO3− in the xylem of crops and Arabidopsis [64,138,139]. NRT1.5, a member of the Nitrate Transporter 1/Peptide Transporter Family (NPF7.3), is important for the NO3−-dependent K+ translocation in Arabidopsis [140-141]. Lack of NRT1.5 resulted in K+ deficiency in shoots under low NO3− availability, whereas the root elemental composition was unchanged [140,141]. Mutant analyses revealed that both NRT1.5 and SKOR contributed additively to K+ translocation; SKOR activity was dominant under high NO3− and low K+ supply, and NRT1.5 was required under low NO3− [142-,43]. Together, these data indicate that NRT1.5 facilitates K+ release out of root parenchyma cells and loading into xylem vessels (Figure 2). NRT1.5 is a plasma membrane protein that in Xenopus oocytes behaved as a low-affinity, pH-dependent bidirectional nitrate transporter [140]. Surprisingly, NRT1.5 has also been shown to release K+ from Xenopus oocytes and yeast in a pH-dependent manner, and has been proposed to function as a K+/H+ antiporter [143]. Recent knowledge gained about the coordinated regulation of K+ and NO3− uptake and nutrition. In fact, K+ starvation is required for triggering high-affinity HAK5-mediated K+ uptake in roots of Arabidopsis and tomato. However, limitation of K+, N, or P, induces hyperpolarization of the plasma membrane of root cells and enhanced HAK5 transcription [144], a response that could be due to maintenance of electrical balance since single N and P starvation, probably resulting in lower NO3− and PO43− contents, and led to a concomitant reduction of the K+ content [137]. Alternatively, the transport of a nutrient could become inhibited if another nutrient is limiting [145]. In line with this, NO3−, PO43−, and SO42− deficiencies reduced root K+ uptake [139]. Furthermore, comparison of the transcriptional responses to single or multiple nutrient deprivations showed that N starvation had a dominant effect over P and K starvation. In other words, the transcriptional landscape of combined K+ and N limitation was mainly driven by the N-starvation response.
The CIPK23/CBL1,9 protein kinase complex is key factor in the coordination of plant nutrition, regulating iron, NO3−, and K+ uptakes [146-149]. The transport and regulatory protein AtNRT1.1 (Nitrate Transporter 1) is involved in both high-affinity and low-affinity nitrate uptake. Unphosphorylated AtNRT1.1 is a low-affinity nitrate transporter working as a dimer, and its phosphorylation by CIPK23/CBL1,9 leads to dimer dissociation. Phosphorylated AtNRT1.1 monomer shows a higher nitrate affinity than the dimers [146,150]. On the other hand, AtAMT1, an ammonium transporter, works as trimers and the phosphorylation by CIPK23/CBL1 (and not CBL9) of a single monomer exhibits an allosteric effect, leading to the cooperative closure of all three pores in the trimer [148]. Together, these data indicate that CIPK23 and CBL1 are major regulators of NO3−, K+, and NH4+ homeostasis in Arabidopsis.
Genetic engineering of specific transporters modifies salinity tolerance
Several obvious ways to achieve salinity tolerance include: (1) decreasing sodium conductance and increasing potassium/sodium selectivity of plasma membrane of root epidermal cells; (2) increasing sodium efflux by root epidermal cells; (3) increasing sodium accumulation in vacuoles; (4) altering sodium and potassium loading and unloading to xylem and phloem depending on plant strategy to cope with salinity. Successful attempts to overexpress or knockout genes of vacuolar proton pump H+-PPase, NHX, HKT, or SOS1-like transporters and to modulate the salinity tolerance of plants had already been reported. Overexpression of the vacuolar H+-PPase would enhance the proton pumping activity at vacuolar membrane and thus permit to accumulate more Na+ in vacuoles due to activity of Na+(cation)/H+ antiporters NHX. The choice of H+-pyrophosphatase is explained by a single gene required for the protein, while the other vacuolar H+-ATPase is composed of several subunits and needs correct overexpression of several genes [80]. Overexpression of vacuolar H+-PPase under control of strong non-specific viral 35S promoter sharply increased salinity tolerance in Arabidopsis, to 250 mM of NaCl [82]. Further attempts to overexpress vacuolar H+-PPases from different plant species increased salinity tolerance in tobacco [151-153].
Other candidates for overexpression are vacuolar NHX genes. Overexpression of AtNHX1 increased salinity tolerance in Arabidopsis to 200 mM NaCl. The overexpressing plants accumulated more Na+ compared to wild type and demonstrated higher Na+/H+ exchange activity in isolated leaf vacuoles [25]. The approach of overexpressing AtNHX1 to improve salinity tolerance proved to be successful for tomato; the transgenic plants accumulated more sodium in leaves but not in fruits at 200 mM NaCl [29]. Cotton plants with AtNHX1 from Arabidopsis [154], rice overexpressing SsNHX1 from halophyte Suaeda salsa [155], tomato with heterologous NHX from Pennisetum glaucum [156] also showed increased salinity tolerance. Overexpression of NHX did not influence the phenotype of plants under control conditions [25,29,153-157]. The results with heterologous expression or overexpression of NHX transporters lead to conclusions that the gene is among determinants and potential candidates for engineering salinity tolerance (e.g., [155,158] with more references for successful overexpression of NHX to increase salinity tolerance in sugar beet, wheat, maize and the other plants). The overexpression of NHX was not tissue-specific and under the control of strong promoters, however, a report could not confirm increase in salinity tolerance in Arabidopsis overexpressing AtNHX1 [74]. Expression in a tissue-specific manner could be the next step for using NHX to increase salinity tolerance.
The amazing simplicity of the idea to play with the expression of known and functionally well characterized transporters and get salt tolerant or salt sensitive plants is applied to plasma membrane SOS1 Na+/H+ antiporters and Na+ or Na+/K+ HKT transporters. SOS1 is expressed in (1) epidermal root cells where it participates in sodium efflux and in (2) xylem parenchyma cells where SOS1 may load Na+ to xylem under moderate salinity and unloads Na+ under high salinity or has more complex mode of xylem loading/unloading [17,18,56,159-161]. Arabidopsis mutants with defects in gene of SOS1 exhibited strong growth inhibition under salt treatment [162], which was rescued in SOS1 mutant by overexpression of SOS1 gene under 35S promoter [56]. Overexpression of SOS1 gene in wild type plants under 35S promoter enhanced salinity tolerance of Arabidopsis at 100–200 mM NaCl [74,163], reduced sodium accumulation in shoots and sodium concentration in xylem sap [160]. Further, overexpression of SOS1 from A. thaliana increased salinity tolerance in transgenic tobacco [75]. SOS1 gene from durum wheat conferred salinity tolerance to SOS1 mutant of Arabidopsis [164]. Interestingly, the effects of overexpression were observed under salt treatment, while in the absence of stress no differences were observed in growth or morphology between wild-type plants and the transgenic lines. Disruption of SOS1 activity by RNA interference in Thellungiella on the opposite resulted in the loss of tolerance of the halophyte indicating importance of Na+ efflux and essential role of SOS1 in salinity tolerance [18]. RNA interference of SOS1 significantly changed the whole transcriptome of Thellungiella [158] and vacuolar pH under salt treatment [67]. A more complicated situation emerges due to tissue-specific expression. SOS1 is important for long-distance ion transport and xylem loading/unloading in Arabidopsis ([17] discussed in: de Boer and Volkov, [165], sodium partioning between plant organs in tomato [161] and ion fluxes in root meristem zone [166], therefore attempts to express it in specific tissues could increase salinity tolerance to a higher extent.
Genetic modification of salinity tolerance using HKT transporters was also successful. Analysis of Arabidopsis plants with mutated HKT gene revealed higher salt sensitivity of the mutants under long term stress, higher sodium accumulation in their shoots under mild salinity treatment [167] and suggested that HKT is involved in recirculation of sodium within plants [48]. Further study confirmed increased sodium in the shoots of Arabidopsis hkt1;1 mutant and clarified that HKT is important for root accumulation of Na+ and Na+ uptake from xylem in Arabidopsis [46]. The next step was to create plants overexpressing HKT [168]. Arabidopsis plants overexpressing AtHKT under the control of 35S promoter were compared with plants specifically overexpressing HKT in cells of root stele. Pro35S:HKT1;1 plants were salt sensitive probably due to higher Na+ uptake by roots while tissue specific overexpression of HKT in stele increased salinity tolerance and reduced sodium accumulation in shoots [168]. The approach was applied to rice where gene from Arabidopsis AtHKT1;1 was heterologously expressed in root cortex. It resulted in lower shoot Na+ concentrations, improved salinity tolerance and involved up- and down-regulation of several membrane transport genes including vacuolar H+-pyrophosphatases [169]. Overexpression of HKT had none [169-171] or slight inhibiting pleiotropic effect on growth without NaCl depending on type of promotor for expression and on plant line studied [168,169]. HKT transporters proved to be important for Na+ exclusion in wheat and were transferred from durum wheat to bread wheat by interspecific crossing; the genes gave beneficial effects including higher K+/Na+ ratio in leaves under saline conditions [21]. Remarkably, the recent introgression of an ancestral form of the HKT1;5 gene from the more Na+-tolerant wheat relative Triticum monococcum into susceptible commercial durum wheat (Triticum turgidum ssp durum) increased grain yields on saline soil by 25% in the field, illustrating the immense potential of this mechanism [23]. Some plants including barley accumulate Na+ in shoots; overexpression of barley HvHKT2;1 under 35S promoter in barley increased salinity tolerance at 100 mM NaCl, but opposite to Arabidopsis increased Na+ concentration in xylem and Na+ accumulation in barley leaves [170]. Taken together the results set HKT transporters to potential candidates for engineering salinity tolerance and among the determinants of the trait (reviewed in: [171-173].
The application of nitrogen (N) fertilizers has greatly increased crop yields. Therefore, enhancing crop nitrogen utilization efficiency is an important goal [174]. For most crops, nitrate is the primary nitrogen source and so enhancing nitrate uptake is one strategy for improving nitrogen utilization efficiency. Multiple nitrate uptake transporters of the NRT1 and NRT2 families work together to enable nitrogen uptake in plants [175,176]. Therefore, nitrate transporters and other proteins that regulate nitrate uptake and sensing provide potential tools for engineering crops with tailored N uptake activity, N metabolism and improved root growth for enhanced nitrogen-use efficiency and reduced-N-fertilizer requirements [177-179].
Although plant salt tolerance at the level of Na+ transport is well characterized, the initial plant perception of salt stress and its transduction to subsequent signaling cascades is still obscure. Many genes targets involved in salt tolerance have been identified through various approaches, particularly through transcriptomics studies. Moreover, it appears that forward genetics and yeast complementation strategies have so far been the most successful approaches to identify relevant targets. The accumulative data show importance of two particular classes of transporters: HKTs which function in both Na+ uptake and long-distance translocation and NHXs in their capacity as H+: Na+ antiport or by maintaining K+ homeostasis. The significance of these systems is often isoform dependent and may be further complicated by allelic variation between cultivars. Manipulation of several of the genes discussed above has been shown to alter uptake, efflux, translocation and compartmentation of Na+. Although in some of these cases improved tolerance can be observed in controlled conditions it has not yet resulted in plants with significantly improved tolerance in field conditions. Simultaneous upregulation of extruding mechanisms through overexpression of systems such as vacuolar pumps, NHXs and SOS1 and loss of function in uptake pathways such as non-selective ion channels and HKTs promises large degrees of additive or synergistic benefits. This is technically challenging but becoming more and more feasible. For instance, salinity tolerance that operates by removal of toxic sodium ions from the xylem sap could be combined with traits that enhance sequestration of sodium into vacuoles, to confer additional salt tolerance. More work will be needed to determine whether or not traits will be compatible when combined. Moreover, many fundamental mechanisms for essential transport processes remain to be uncovered and many essential transporters undoubtedly remain to be discovered. Therefore, knowledge-targeted pyramiding of traits will require future advances in fundamental research into plant membrane transport processes.
This work was supported through funding by the Ministry of Higher Education and Scientific Research of Tunisia.
- Shrivastava P, Kumar R. Soil salinity: A serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci. 2015; 22: 123-131. PubMed: https://pubmed.ncbi.nlm.nih.gov/25737642/
- Shahbaz M, Ashraf M. Improving salinity tolerance in cereals. Crit. Rev. Plant Sci. 2013; 32: 237–249.
- Munns R. Genes and salt tolerance: bringing them together. New Phytol. 2005; 167: 645–663. PubMed: https://pubmed.ncbi.nlm.nih.gov/16101905/
- Jamil A, Riaz S, Ashraf M, Foolad MR. Gene expression profiling of plants under salt stress. Crit Rev Plant Sci. 2011; 30: 435–458.
- van Ittersum MK, van Bussel LGJ, Wolf J, Grassini P, et al. Can sub-Saharan Africa feed itself? Proc. Natl Acad Sci. U.S.A. 2016; 113: 14964–14969. PubMed: https://pubmed.ncbi.nlm.nih.gov/27956604/
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008; 59: 651–681. PubMed: https://pubmed.ncbi.nlm.nih.gov/18444910/
- Cramer GR, Läuchli A, Polito VS. Displacement of Ca2+ by Na+ from the plasmalemma of root cells: a primary response to salt stress? Plant Physiol. 1985; 79: 207–211. PubMed: https://pubmed.ncbi.nlm.nih.gov/16664372/
- Kinraide TB. Interactions among Ca2+, Na+ and K+ in salinity toxicity: quantitative resolution of multiple toxic and ameliorative effects. J Exp Bot. 1999; 50: 1495–1505.
- Zhu JK. Regulation of ion homeostasis under salt stress. Curr Opin Plant Biol. 2003; 6: 441–445. PubMed: https://pubmed.ncbi.nlm.nih.gov/12972044/
- Marin K, Suzuki I, Yamaguchi K, Ribbeck K, Yamamoto H, et al. Identification of histidine kinases that act as sensors in the perception of salt stress in Synechocystis sp. PCC 6803. Proc Natl Acad Sci. U.S.A. 2003; 100: 9061–9066. PubMed: https://pubmed.ncbi.nlm.nih.gov/12853569/
- Tran LSP, Urao T, Qin F, Maruyama K, Kakimoto T, et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci. U.S.A. 2007; 104: 20623–20628. PubMed: https://pubmed.ncbi.nlm.nih.gov/18077346/
- Shabala S, Wu H, Bose J. Salt stress sensing and early signaling events in plant roots: current knowledge and hypothesis. Plant Sci. 2015; 109–119. PubMed: https://pubmed.ncbi.nlm.nih.gov/26706063/
- Sun J, Zhang X, Deng S, Zhang C, Wang M, et al. Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS One 2012; 7: e53136.
- Wu H, Shabala L, Liu X, Azzarello E, Zhou M, et al. Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front Plant Sci. 2015; 6: 71.
- Wu H. Tissue Specificity of Cytosolic K+ Retention, Na+ Extrusion, and Vacuolar Na+ Sequestration Traits in the Context of Differential Salinity Stress Tolerance in Barley and Wheat (Ph.D. Dissertation) University of Tasmania, Australia. 2015. PubMed: https://eprints.utas.edu.au/23053/
- Apse MP, Blumwald E. Na+ transport in plants. FEBS Lett. 2007; 581: 2247–2254. PubMed: https://pubmed.ncbi.nlm.nih.gov/17459382/
- Shi H, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1controls long-distance Na+ transport in plants. Plant Cell 2002; 14: 465–477. PubMed: https://pubmed.ncbi.nlm.nih.gov/11884687/
- Oh DH, Leidi E, Zhang Q, Hwang SM, Li Y, et al. Loss of halophytism by interference with SOS1 expression. Plant Physiol. 2009a; 151: 210–222. PubMed: https://pubmed.ncbi.nlm.nih.gov/19571313/
- Al-Karaki GN. Growth, water use efficiency, and sodium and potassium acquisition by tomato cultivars grown under salt stress. J Plant Nutr. 2000; 23: 1–8.
- Byrt CS, Platten JD, Spielmeyer W, James RA, Lagudah ES, et al. HKT1;5-like cation transporters linked to Na+ exclusion loci in wheat, Nax2 and Kna1. Plant Physiol. 2007; 143: 1918–1928. PubMed: https://pubmed.ncbi.nlm.nih.gov/17322337/
- James RA, Blake C, Byrt CS, Munns R. Major genes for Na+ exclusion, Nax1 and Nax2 (wheat HKT1;4 and HKT1;5), decrease Na+ accumulation in bread wheat leaves under saline and water-logged conditions. J Exp Bot. 2011; 62: 2939–2947. PubMed: https://pubmed.ncbi.nlm.nih.gov/21357768/
- Møller IS, Tester M. Salinity tolerance of Arabidopsis: A good model for cereals? Trends Plant Sci. 2007; 12: 534–540. PubMed: https://pubmed.ncbi.nlm.nih.gov/18023242/
- Munns R, James RA, Xu B, Athman A, Conn SJ, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol. 2012; 30: 360–364. PubMed: https://pubmed.ncbi.nlm.nih.gov/22407351/
- Roy SJ, Huang W, Wang XJ, Evrard A, Schmöckel SM, et al. A novel protein kinase involved in Na+ exclusion revealed from positional cloning. Plant Cell Environ. 2013; 36: 553–568. PubMed: https://pubmed.ncbi.nlm.nih.gov/22897323/
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Sci. 1999; 285: 1256–1258. PubMed: https://pubmed.ncbi.nlm.nih.gov/10455050/
- Mansour MMF, Salama KHA, Al-Mutawa MM. Transport proteins and salt tolerance in plants. Plant Sci. 2003; 164: 891–900.
- Rahnama A, Poustini K, Tavakkol-Afshari R, Ahmadi A, Alizadeh H. Growth properties and ion distribution in different tissues of bread wheat genotypes (Triticum aestivum L.) differing in salt tolerance. J Agron Crop Sci. 2011; 197; 21–30.
- Maathuis FJM, Amtmann A. K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot. 1999; 84: 123–133.
- Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol. 2001; 19: 765–768. PubMed: https://pubmed.ncbi.nlm.nih.gov/11479571/
- Chen H, An R, Tang JH, Cui XH, Hao FS, et al. Over-expression of a vacuolar Na+/H+ antiporter gene improves salt tolerance in an upland rice. Mol Breed. 2007; 19: 215–225.
- Gouiaa S, Khoudi H, Leidi EO, Pardo JM, Masmoudi K. Expression of wheat Na+/H+ antiporter TNHXS1 and H+-pyrophosphatase TVP1 genes in tobacco from a bicistronic transcriptional unit improves salt tolerance. Plant Mol Biol. 2012; 79: 137–155. PubMed: https://pubmed.ncbi.nlm.nih.gov/22415161/
- Cuin TA, BOSE J, Stefano G, JHA D, Tester M, et al. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant Cell Environ. 2011; 34: 947–961. PubMed: https://pubmed.ncbi.nlm.nih.gov/21342209/
- Fukuda A, Nakamura A, Tagiri A, Tanaka H, Miyao A, et al. Function, intracellular localization and the importance in salt tolerance of a vacuolar Na+/H+ antiporter from rice. Plant Cell Physiol. 2004; 45: 146–159. PubMed: https://pubmed.ncbi.nlm.nih.gov/14988485/
- Bonales-Alatorre E, Shabala S, Chen ZH, Pottosin I. Reduced tonoplast fast-activating and slow-activating channel activity is essential for conferring salinity tolerance in a facultative halophyte, quinoa. Plant Physiol. 2013; 162: 940–952. PubMed: https://pubmed.ncbi.nlm.nih.gov/23624857/
- Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot. 2013; 112: 1209–1221. PubMed: https://pubmed.ncbi.nlm.nih.gov/24085482/
- Maathuis FJM. Sodium in plants: perception, signaling, and regulation of sodium fluxes. J Exp Bot. 2014; 65: 849–858. PubMed: https://pubmed.ncbi.nlm.nih.gov/24151301/
- Colmenero-Flores JM, Martínez G, Gamba G, Vázquez N, Iglesias DJ, et al. Identification and functional characterization of cation-chloride co-transporters in plants. Plant J. 2007; 50: 278–292. PubMed: https://pubmed.ncbi.nlm.nih.gov/17355435/
- Wegner LH, De Boer AH. Two inward K+ channels in the xylem parenchyma cells of barley roots are regulated by G-protein modulators through a membrane-delimited pathway. Planta. 1997; 203: 506–516.
- Yadav N, Shukla P, Jha A, Agarwal PK, Jha B. The SbSOS1 gene from the extreme halophyte Salicornia brachiate enhances Na+ loading in xylem and confers salt tolerance in transgenic tobacco. BMC Plant Biol. 2012; 12: 188. PubMed: https://pubmed.ncbi.nlm.nih.gov/23057782/
- Zhu M, Shabala L, Cuin TA, Huang X, Zhou M, et al. Nax loci affect SOS1-like Na+/H+ exchanger expression and activity in wheat. J Exp Bot. 2016; 67: 835–844. PubMed: https://pubmed.ncbi.nlm.nih.gov/26585227/
- Sunarpi, Horie T, Motoda J, Kubo M, Yang H, et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na+ unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005; 44: 928–938. PubMed: https://pubmed.ncbi.nlm.nih.gov/16359386/
- Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, et al. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol. 2006; 142: 1718–1727. PubMed: https://pubmed.ncbi.nlm.nih.gov/17071645/
- Jaime-Pérez N, Pineda B, García-Sogo B, Atares A, Athman A, et al. The sodium transporter encoded by the HKT1;2 gene modulates sodium/potassium homeostasis in tomato shoots under salinity. Plant Cell Environ. 2017; 40: 658–671. PubMed: https://pubmed.ncbi.nlm.nih.gov/27987209/
- Kong X, Luo Z, Dong H, Eneji AE, Li W. Effects of non-uniform root zone salinity on water use, Na+ recirculation, and Na+ and H+ flux in cotton. J Exp Bot. 2012; 63: 2105–2116. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3295398/
- Rus A, Lee BH, Muñoz-Mayor A, et al. AtHKT1 Facilitates Na+ Homeostasis and K+ Nutrition in Planta. Plant Physiol. 2004; 136: 2500-2511. PubMed: https://pubmed.ncbi.nlm.nih.gov/15347798/
- Davenport RJ, Muñoz-Mayor A, Jha D, Essah PA, Rus A, et al. The Na+ transporter AtHKT1;1 control retrieval of Na+ from the xylem in Arabidopsis. Plant Cell Environ. 207; 30: 497–507. PubMed: https://pubmed.ncbi.nlm.nih.gov/17324235/
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003; 91: 503–527. PubMed: https://pubmed.ncbi.nlm.nih.gov/12646496/
- Berthomieu P, Conéjéro G, Nublat A, Brackenbury WJ, Lambert C, et al. Functional analysis of AtHKT1 in Arabidopsis shows that Na+ recirculation by the phloem is crucial for salt tolerance, EMBO J. 2003; 22: 2004–2014. PubMed: https://pubmed.ncbi.nlm.nih.gov/12727868/
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005; 37: 1141–1146. PubMed: https://pubmed.ncbi.nlm.nih.gov/16155566/
- Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, et al. OsHKT1;5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J. 2017; 91: 657–670. PubMed: https://pubmed.ncbi.nlm.nih.gov/28488420/
- Wang Y, Wu WH. Potassium transport and signaling in higher plants, Annu. Rev. Plant Biol. 2013; 64: 451–476. PubMed: https://pubmed.ncbi.nlm.nih.gov/23330792/
- Shi H, Ishitani M, Kim C, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc Natl Acad Sci. U.S.A. 2000; 97: 6896–6901. PubMed: https://pubmed.ncbi.nlm.nih.gov/10823923/
- Shabala S, Shabala L, Van Volkenburgh E, Newman I. Effect of divalent cations on ion fluxes and leaf photochemistry in salinized barley leaves. J Exp Bot. 2005; 56: 1369–1378. PubMed: https://pubmed.ncbi.nlm.nih.gov/15809285/
- Liu J, Ishitani M, Halfter U, Kim CS, Zhu JK. The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc Natl Acad Sci. U.S.A. 2000; 97: 3730–3734. PubMed: https://pubmed.ncbi.nlm.nih.gov/10725382/
- Luan S, Lan W, Lee SC. Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL-CIPK network. Curr Opin. Plant Biol. 2009; 12: 339–346. PubMed: https://pubmed.ncbi.nlm.nih.gov/19501014/
- Halfter U, Ishitani M, Zhu JK. The Arabidopsis SOS2 protein kinase physically interacts with and is activated by the calcium-binding protein SOS3. Proc. Natl Acad Sci. U.S.A. 2000; 97: 3735–3740. PubMed: https://pubmed.ncbi.nlm.nih.gov/10725350/
- Shono M, Wada M, Hara Y, Fujii T. Molecular cloning of Na+-ATPase cDNA from a marine alga, Heterosigma akashiwo. Biochim. Biophys. Acta Biomembr. 2001; 1511: 193–199. PubMed: https://pubmed.ncbi.nlm.nih.gov/11248217/
- Lunde C, Drew DP, Jacobs AK, Tester M. Exclusion of Na+ via sodium ATPase (PpENA1) ensures normal growth of Physcomitrella patens under moderate salt stress. Plant Physiol. 2007; 144: 1786–1796. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1949878/
- Demidchik V, Maathuis FJM. Physiological roles of nonselective cation channels in plants: from salt stress to signaling and development. New Phytol. 2007; 175: 387–404.
- Zhang JL, Flowers TJ, Wang SM. Mechanisms of sodium uptake by roots of higher plants. Plant Soil. 2010; 326: 45–60.
- Demidchik V, Tester M. Sodium fluxes through nonselective cation channels in the plasma membrane of protoplasts from Arabidopsis roots. Plant Physiol. 2002; 128: 379–387. PubMed: https://pubmed.ncbi.nlm.nih.gov/11842142/
- Maathuis FJ, Sanders D. Sodium uptake in Arabidopsis roots is regulated by cyclic nucleotides. Plant Physiol. 2001; 127: 1617–1625. PubMed: https://pubmed.ncbi.nlm.nih.gov/11743106/
- Oh DH, Zahir A, Yun DJ, Bressan RA, Bohnert HJ. SOS1 and halophytism. Plant Signal Behav. 2009; 4: 1081–1083. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2819520/
- Quan R, Wang J, Yang D, Zhang H, Zhang Z, et al. EIN3 and SOS2 synergistically modulate plant salt tolerance. Sci. Rep. 2017; 7: 44637. PubMed: https://pubmed.ncbi.nlm.nih.gov/28300216/
- Gong D, Guo Y, Schumaker KS, Zhu JK. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiol. 2004; 134: 919–926. PubMed: https://pubmed.ncbi.nlm.nih.gov/15020756/
- Bose J, Rodrigo-Moreno A, Lai D, Xie Y, Shen W, et al. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann Bot. 2015; 115: 481-494. PubMed: https://pubmed.ncbi.nlm.nih.gov/25471095/
- Chung JS, Zhu JK, Bressan RA, Hasegawa PM, Shi H. Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 2008; 53: 554–565. PubMed: https://pubmed.ncbi.nlm.nih.gov/17996020/
- Katiyar-Agarwal S, Zhu JJ, Kim K, Agarwal M, Fu X, et al. The plasma membrane Na+/H+ antiporter SOS1 interacts with RCD1 and functions in oxidative stress tolerance in Arabidopsis. Proc Natl Acad Sci. U.S.A. 2006; 103: 18816–18821. PubMed: https://pubmed.ncbi.nlm.nih.gov/17023541/
- Sagor GHM, Zhang S, Kojima S, Simm S, Berberich T, et al. Reducing cytoplasmic polyamine oxidase activity in Arabidopsis increases salt and drought tolerance by reducing reactive oxygen species production and increasing defense gene expression. Front Plant Sci. 2016; 7: 214. PubMed: https://pubmed.ncbi.nlm.nih.gov/26973665/
- Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, et al. Overexpression of SOS (salt overly sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant. 2009; 2: 22–31. PubMed: https://pubmed.ncbi.nlm.nih.gov/19529826/
- Yue Y, Zhang M, Zhang J, Duan L, Li Z. SOS1 gene overexpression increased salt tolerance in transgenic tobacco by maintaining a higher K+/Na+ ratio. J Plant Physiol. 2012; 169: 255–261. PubMed: https://pubmed.ncbi.nlm.nih.gov/22115741/
- Leidi EO, Barragán V, Rubio L, El-Hamdaoui A, Ruiz MT, et al. The AtNHX1 exchanger mediates potassium compartmentation in vacuoles of transgenic tomato. Plant J. 2010; 61: 495–506.
- Bassil E, Tajima H, Liang YC, Ohto MA, Ushijima K, et al. The Arabidopsis Na+/H+ antiporters NHX1 and NHX2 control vacuolar pH and K+ homeostasis to regulate growth, flower development, and reproduction. Plant Cell. 2011; 23: 3482–3497. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3203450/
- Barragan V, Leidi EO, Andres Z, Rubio L, De Luca A, et al. Ion exchangers NHX1 and NHX2 mediate active potassium uptake into vacuoles to regulate cell turgor and stomatal function in Arabidopsis. Plant Cell. 2012; 24: 1127–1142. PubMed: https://pubmed.ncbi.nlm.nih.gov/22438021/
- Liu X, Cai S, Wang G, Wang F, Dong F, et al. Halophytic NHXs confer salt tolerance by altering cytosolic and vacuolar K+ and Na+ in Arabidopsis root cell. Plant Growth Regul. 2017; 82: 333–351.
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, et al. The Arabidopsis thaliana proton transporters, AtNhx1 and Avp1, can function in cation detoxification in yeast. Proc Natl Acad Sci. U.S.A. 1999; 96: 1480–1485. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC15488/
- Adem G, Roy SJ, Zhou M, Bowman JP, Shabala S. Evaluating contribution of ionic, osmotic and oxidative stress components towards salinity tolerance in barley. BMC Plant Biol. 2014; 14: 113. PubMed: https://pubmed.ncbi.nlm.nih.gov/24774965/
- Sandhu D, Cornacchione MV, Ferreira JFS, Suarez DL. Variable salinity responses of 12 alfalfa genotypes and comparative expression analyses of salt-response genes. Sci Rep. 2017; 7: 42958. PubMed: https://pubmed.ncbi.nlm.nih.gov/28225027/
- Mullan DJ, Colmer TD, Francki MG. Arabidopsis-rice-wheat gene orthologues for Na+ transport and transcript analysis in wheat-L. elongatum aneuploids under salt stress. Mol Gen Genomics. 2007; 277: 199–212. PubMed: https://pubmed.ncbi.nlm.nih.gov/17103227/
- Silva P, Gerós H. Regulation by salt of vacuolar H+-ATPase and H+-pyrophosphatase activities and Na+/H+ exchange. Plant Signal. Behav. 2009; 4: 718–726. PubMed: https://pubmed.ncbi.nlm.nih.gov/19820346/
- Baisakh N, Ramanarao MV, Rajasekaran K, Subudhi P, Janda J, et al. Enhanced salt stress tolerance of rice plants expressing a vacuolar H+-ATPase subunit c1 (SaVHAc1) gene from the halophyte grass Spartina alterniflora Löisel. Plant Biotechnol J. 2012; 10: 453–464. PubMed: https://pubmed.ncbi.nlm.nih.gov/22284568/
- Gaxiola RA, Li J, Undurraga S, Dang LM, Allen GJ, et al. Drought- and salt-tolerant plants result from overexpression of the AVP1 H+-pump. Proc Natl Acad Sci. U.S.A. 2001; 98: 11444–11449. PubMed: https://pubmed.ncbi.nlm.nih.gov/11572991/
- Qui QS, Guo Y, Quintero FJ, Pardo JM, Schumaker KS, et al. Regulation of vacuolar Na+/H+ exchange in Arabidopsis thaliana by the Salt-Overly-Sensitive (SOS) pathway. J Biol Chem. 2004; 279: 207–215. PubMed: https://pubmed.ncbi.nlm.nih.gov/14570921/
- Yamaguchi T, Aharon GS, Sottosanto JB, Blumwald E. Vacuolar Na+/H+ antiporter cation selectivity is regulated by calmodulin from within the vacuole in a Ca2+- and pH-dependent manner. Proc Natl Acad Sci. U.S.A. 2005; 102: 16107–16112. PubMed: https://pubmed.ncbi.nlm.nih.gov/16249341/
- Quan R, Lin H, Mendoza I, Zhang Y, Cao W, et al. SCABP8/CBL10, a putative calcium sensor, interacts with the protein kinase SOS2 to protect Arabidopsis shoots from salt stress. Plant Cell. 2007; 19: 1415–1431. PubMed: https://pubmed.ncbi.nlm.nih.gov/17449811/
- Tang RJ, Yang Y, Yang L, Liu H, Wang CT, et al. Poplar calcineurin B-like proteins PtCBL10A and PtCBL10B regulate shoot salt tolerance through interaction with PtSOS2 in the vacuolar membrane. Plant Cell Environ. 2014; 37: 573–588. PubMed: https://pubmed.ncbi.nlm.nih.gov/23941462/
- Kim BG, Waadt R, Cheong YH, Pandey GK, Dominguez-Solis JR, et al. The calcium sensor CBL10 mediates salt tolerance by regulating ion homeostasis in Arabidopsis. Plant J. 2007; 52: 473–484. PubMed: https://pubmed.ncbi.nlm.nih.gov/17825054/
- Assaha DVM, Ueda A, Saneoka H, Al-Yahyai R, Yaish MW. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017; 8: 509. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5513949/
- White PG, Broadley MR. Chloride in soils and its uptake and movement with the plant. Ann Bot. 2001; 88: 967-988.
- Flowers TJ, Colmer TD. Salinity tolerance in halophytes. New Phytol. 2008; 179: 945-963.
- Hechenberger M, Schwappah B, Fischer WN, Frommer WB, Jentsch TJ, et al. A family of putative chloride channels from Arabidopsis and functional complementation of a yeast strain with a CLC gene distruption. J Biol Chem. 1996; 271: 33632-33638. PubMed: https://pubmed.ncbi.nlm.nih.gov/8969232/
- Diédhiou CJ, Golldack D. Salt-dependent regulation of chloride channel transcripts in rice. Plant Sci. 2006; 170: 793-800.
- Xu G, Magen H, Tarchitzky J, Kafkafi U. Advances in chloride nutrition of plants. Adv. Agronom. 2000; 68: 97-150.
- De Angeli A, Thomine S, Franchisse JM, Ephritikhinea G, Gambale F, et al. Anions channels and transporters in plant cell membranes. FEBS Lett. 2007; 581: 2367-2374. PubMed: https://pubmed.ncbi.nlm.nih.gov/17434490/
- Diédhiou CJ. Mechanisms of salt tolerance: sodium, chloride and potassium homeostasis in two rice lines with different tolerance to salinity stress. PhD thesis 2006; University of Bielefeld, Germany.
- Li WYF, Wong FL, Tsai SN, Phang TH, Shao G, et al. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 2006; 29: 1122-1137. PubMed: https://pubmed.ncbi.nlm.nih.gov/17080938/
- Nakamura A, Fukuda A, Sakai S, Tanaka Y. Molecular cloning, functional expression and subcellular localization of two putative vacuolar voltage-gated chloride channels in rice (Oryza sativa L.). Plant Cell Physiol. 2006; 47: 32-42. PubMed: https://pubmed.ncbi.nlm.nih.gov/16249326/
- Hedrich R. Ion channels in plants. Physiol Rev. 2012; 92: 1777–1811. PubMed: https://pubmed.ncbi.nlm.nih.gov/23073631/
- Nieves-Cordones M, Aleman F, Martinez V, Rubio F. K+ uptake in plant roots. The systems involved, their regulation and parallels in other organisms. J Plant Physiol. 2014; 171: 688–695. PubMed: https://pubmed.ncbi.nlm.nih.gov/24810767/
- Li W, Xu G, Alli A, Yu. Plant HAK/KUP/KT K+ transporters: function and regulation. Semin Cell Dev. Biol. 2018; 74: 133–141. PubMed: https://pubmed.ncbi.nlm.nih.gov/28711523/
- Hamamoto S, Horie T, Hauser F, Deinlein U, Schroeder JI, et al. HKT transporters mediate salt stress resistance in plants: from structure and function to the field. Curr Opin Biotechnol. 2015; 32: 113–120. PubMed: https://pubmed.ncbi.nlm.nih.gov/25528276/
- Sze H, Chanroj S. Plant endomembrane dynamics: studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiol. 2018; 177: 875–895. PubMed: https://pubmed.ncbi.nlm.nih.gov/29691301/
- Hirsch RE, Lewis BD, Spalding EP, Sussman MR. A role for the AKT1 potassium channel in plant nutrition. Sci. 1998; 280: 918–921. PubMed: https://pubmed.ncbi.nlm.nih.gov/9572739/
- Dreyer I, Uozumi N. Potassium channels in plant cells. FEBS J. 2011; 278: 4293–4303. PubMed: https://pubmed.ncbi.nlm.nih.gov/21955642/
- Jeanguenin L, Alcon C, Duby G, Boeglin M, Cherel I, et al. AtKC1 is a general modulator of Arabidopsis inward Shaker channel activity. Plant J. 2011; 67: 570–582. PubMed: https://pubmed.ncbi.nlm.nih.gov/21518051/
- Gobert A, Isayenkov S, Voelker C, Czempinski K, Maathuis FJ. The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proc Natl Acad Sci. U.S.A. 2007; 104: 10726–10731. PubMed: https://pubmed.ncbi.nlm.nih.gov/17563365/
- Latz A, Mehlmer N, Zapf S, Mueller TD, Wurzinger B, et al. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol Plant. 2013; 6: 1274–1289. PubMed: https://pubmed.ncbi.nlm.nih.gov/23253603/
- Peiter E, Maathuis FJ, Mills LN, Knight H, Pelloux J, et al. The vacuolar Ca2+-activated channel TPC1 regulates germination and stomatal movement. Nature. 2005; 434: 404–408. PubMed: https://pubmed.ncbi.nlm.nih.gov/15772667/
- Hedrich R, Mueller TD, Becker D, Marten I. Structure and function of TPC1 vacuole SV channel gains shape. Mol. Plant 2018; 11: 764–775. PubMed: https://pubmed.ncbi.nlm.nih.gov/29614320/
- Choi WG, Toyota M, Kim SH, Hilleary R, Gilroy S. Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc Natl Acad Sci. U.S.A. 2014; 111: 6497–6502. PubMed: https://pubmed.ncbi.nlm.nih.gov/24706854/
- Evans MJ, Choi WG, Gilroy S, Morris RJ. A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 2016; 171: 1771–1784. PubMed: https://pubmed.ncbi.nlm.nih.gov/27261066/
- Greiner T, Ramos J, Alvarez MC, Gurnon JR, Kang M, et al. Functional HAK/KUP/KT-like potassium transporter encoded by chlorella viruses. Plant J. 2011; 68: 977–986. PubMed: https://pubmed.ncbi.nlm.nih.gov/21848655/
- Santa-Maria GE, Oliferuk S, Moriconi, JI. KT-HAK-KUP transporters in major terrestrial photosynthetic organisms: a twenty years tale. J Plant Physiol. 2018; 226: 77–90. PubMed: https://pubmed.ncbi.nlm.nih.gov/29704646/
- Very AA, Nieves-Cordones M, Daly M, Khan I, Fizames C, et al. Molecular biology of K+ transport across the plant cell membrane: what do we learn from comparison between plant species? J Plant Physiol. 2014; 171: 748–769. PubMed: https://pubmed.ncbi.nlm.nih.gov/24666983/
- Benito B, Haro R, Amtmann A, Cuin TA, Dreyer I. The twins K+ and Na+ in plants. J Plant Physiol. 2014; 171: 723–731. PubMed: https://pubmed.ncbi.nlm.nih.gov/24810769/
- Platten JD, Cotsaftis O, Berthomieu P, Bohnert H, Davenport RJ, et al. Nomenclature for HKT transporters, key determinants of plant salinity tolerance. Trends Plant Sci. 2006; 11: 372–374. PubMed: https://pubmed.ncbi.nlm.nih.gov/16809061/
- Ma YC, Auge RM, Dong C, Cheng ZM. Increased salt tolerance with overexpression of cation/proton antiporter 1 genes: a meta-analysis. Plant Biotechnol J. 2017; 15: 162–173. PubMed: https://pubmed.ncbi.nlm.nih.gov/27383431/
- De Luca A, Pardo JM, Leidi EO. Pleiotropic effects of enhancing vacuolar K/H exchange in tomato. Physiol Plant. 2018; 163: 88–102. PubMed: https://pubmed.ncbi.nlm.nih.gov/29076168/
- Jiang XY, Leidi EO, Pardo JM. How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal Behav. 2010; 5: 792–795. PubMed: https://pubmed.ncbi.nlm.nih.gov/20495345/
- Andrés Z, Pérez-Hormaeche J, Leidi EO, Schlücking K, Steinhorst L, et al. Control of vacuolar dynamics and regulation of stomatal aperture by tonoplast potassium uptake. Proc Natl Acad Sci. U.S.A. 2014; 111: E1806–E1814. PubMed: https://pubmed.ncbi.nlm.nih.gov/24733919/
- Ahmad I, Maathuis FJ. Cellular and tissue distribution of potassium: physiological relevance, mechanisms and regulation. J Plant Physiol. 2014; 171: 708–714. PubMed: https://pubmed.ncbi.nlm.nih.gov/24810768/
- Yang T, Zhang S, Hu Y, Wu F, Hu Q, et al. The role of a potassium transporter OsHAK5 in potassium acquisition and transport from roots to shoots in rice at low potassium supply levels. Plant Physiol. 2014; 166: 945–959. PubMed: https://pubmed.ncbi.nlm.nih.gov/25157029/
- Gaymard F, Pilot G, Lacombe B, Bouchez D, Bruneau D, et al, Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell. 1998; 94: 647–655. PubMed: https://pubmed.ncbi.nlm.nih.gov/9741629/
- Johansson I, Wulfetange K, Porée F, Michard E, Gajdanowicz P, et al. External K+ modulates the activity of the Arabidopsis potassium channel SKOR via an unusual mechanism. Plant J. 2006; 46: 269–281. PubMed: https://pubmed.ncbi.nlm.nih.gov/16623889/
- Thompson MV, Zwieniecki MA. “The role of potassium in long distance transport in plants” in Vascular transport in plants. Eds. NM. Holbrook and MA. Zwieniecki (Burlington: Academic Press). 2005; 221–240.
- De Schepper V, De Swaef T, Bauweraerts I, Steppe K. Phloem transport: a review of mechanisms and controls. J Exp Bot. 2013; 64: 4839–4850. PubMed: https://pubmed.ncbi.nlm.nih.gov/24106290/
- Deeken R, Sanders C, Ache P, Hedrich R. Developmental and light-dependent regulation of a phloem-localised K+ channel of Arabidopsis thaliana. Plant J. 2000; 23: 285–290. PubMed: https://pubmed.ncbi.nlm.nih.gov/10929122/
- Lacombe B, Pilot G, Michard E, Gaymard F, Sentenac H, et al. A Shaker-like K(+) channel with weak rectification is expressed in both source and sink phloem tissues of Arabidopsis. Plant Cell. 2000; 12: 837–851. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC149088/
- Gajdanowicz P, Michard E, Sandmann M, Rocha M, Correa LG, et al. Potassium (K+) gradients serve as a mobile energy source in plant vascular tissues. Proc Natl Acad Sci. U.S.A. 2011; 108: 864–869.
- Dreyer I, Michard E, Lacombe B, Thibaud JB. A plant Shaker-like K+ channel switches between two distinct gating modes resulting in either inward-rectifying or ‘leak’ current. FEBS Lett. 2001; 505: 233–239.
- Chérel I, Lefoulon C, Boeglin M, Sentenac H. Molecular mechanisms involved in plant adaptation to low K+ availability. J Exp Bot. 2014; 65: 833-848. PubMed: https://pubmed.ncbi.nlm.nih.gov/24293613/
- Cherel I, Michard E, Platet N, Mouline K, Alcon C, et al. Physical and functional interaction of the Arabidopsis K+ channel AKT2 and phosphatase AtPP2CA. Plant Cell. 2002; 14: 1133–1146. PubMed: https://pubmed.ncbi.nlm.nih.gov/12034902/
- Michard E, Lacombe B, Porée F, Mueller-Roeber B, Sentenac H, et al. A unique voltage sensor sensitizes the potassium channel AKT2 to phosphoregulation. J Gen Physiol. 2005a; 126: 605–617. PubMed: https://pubmed.ncbi.nlm.nih.gov/16316977/
- Michard E, Dreyer I, Lacombe B, Sentenac H, Thibaud JB. Inward rectification of the AKT2 channel abolished by voltage-dependent phosphorylation. Plant J. 2005b; 44: 783–797. PubMed: https://pubmed.ncbi.nlm.nih.gov/16297070/
- Han M, Wu W, Wu WH, Wang Y. Potassium transporter KUP7 Is involved in K+ acquisition and translocation in Arabidopsis root under K+-limited conditions. Mol Plant. 2016; 9: 437–446. PubMed: https://pubmed.ncbi.nlm.nih.gov/26851373/
- Zhang M, Cao Y, Wang Z, Wang ZQ, Shi J, et al. A retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018; 217: 1161–1176. PubMed: https://pubmed.ncbi.nlm.nih.gov/29139111/
- Kellermeier F, Armengaud P, Seditas TJ, Danku J, Salt DE, et al. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell. 2014; 26: 1480–1496. PubMed: https://pubmed.ncbi.nlm.nih.gov/24692421/
- Engels C, Marschner H. Influence of the form of nitrogen supply on root uptake and translocation of cations in the xylem exudate of maize (Zea mays L). J Exp Bot. 1993; 44: 1695–1701.
- Rodenas R, Garcia-Legaz MF, Lopez-Gomez E, Martinez V, Rubio F, et al. NO3−, PO43− and SO42− deprivation reduced LKT1-mediated low-affinity K+ uptake and SKOR-mediated K(+) translocation in tomato and Arabidopsis plants. Physiol Plant. 2017; 160: 410–424. PubMed: https://pubmed.ncbi.nlm.nih.gov/28244226/
- Lin SH, Kuo HF, Canivenc G, Lin CS, Lepetit M, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell. 2008; 20: 2514–2528. PubMed: https://pubmed.ncbi.nlm.nih.gov/18780802/
- Drechsler N, Zheng Y, Bohner A, Nobmann B, von Wiren N, et al. Nitrate-dependent control of shoot K homeostasis by the nitrate transporter1/peptide transporter family member NPF7.3/NRT1.5 and the stelar K+ outward rectifier SKOR in Arabidopsis. Plant Physiol. 2015; 169: 2832–2847. PubMed: https://pubmed.ncbi.nlm.nih.gov/26508776/
- Meng S, Peng JS, He YN, Zhang GB, Yi HY, et al. Arabidopsis NRT1.5 mediates the suppression of nitrate starvation-induced leaf senescence by modulating foliar potassium level. Mol Plant. 2016; 9: 461–470. PubMed: https://pubmed.ncbi.nlm.nih.gov/26732494/
- Li H, Yu M, Du XQ, Wang ZF, Wu WH, et al. NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell. 2017; 29: 2016–2026. PubMed: https://pubmed.ncbi.nlm.nih.gov/28739644/
- Rubio F, Fon M, Rodenas R, Nieves-Cordones M, Aleman F, et al. A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiol Plant. 2014; 152: 558–570. PubMed: https://pubmed.ncbi.nlm.nih.gov/24716623/
- Nieves-Cordones M, Rodenas R, Lara A, Martinez V, Rubio F. The combination of K(+) deficiency with other environmental stresses: what is the outcome? Physiol Plant. 2019; 165: 264–276. PubMed: https://pubmed.ncbi.nlm.nih.gov/30187486/
- Ho CH, Lin SH, Hu HC, Tsay YF. CHL1 functions as a nitrate sensor in plants. Cell. 2009; 138: 1184–1194. PubMed: https://pubmed.ncbi.nlm.nih.gov/19766570/
- Ragel P, Rodenas R, Garcia-Martin E, Andres Z, Villalta I, et al. The CBL-interacting protein kinase CIPK23 regulates HAK5-mediated high-affinity K+ uptake in Arabidopsis roots. Plant Physiol. 2015; 169: 2863–2873. PubMed: https://pubmed.ncbi.nlm.nih.gov/26474642/
- Straub T, Ludewig U, Neuhauser B. The Kinase CIPK23 inhibits ammonium transport in Arabidopsis thaliana. Plant Cell. 2017; 29: 409–422. PubMed: https://pubmed.ncbi.nlm.nih.gov/28188265/
- Dubeaux G, Neveu J, Zelazny E, Vert G. Metal sensing by the IRT1 transporter-receptor orchestrates its own degradation and plant metal nutrition. Mol Cell. 2018; 69: 953–964.e955. PubMed: https://pubmed.ncbi.nlm.nih.gov/29547723/
- Sun J, Bankston JR, Payandeh J, Hinds TR, Zagotta WN, et al. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature. 2014; 507, 73–77. PubMed: https://pubmed.ncbi.nlm.nih.gov/24572362/
- D’yakova EV, Rakitin AL, Kamionskaya AM, Baikov AA, Lahti R, et al. A study of the effect of expression of the gene encoding the membrane H+-pyrophosphatase of Rhodospirillum rubrum on salt resistance of transgenic tobacco plants. Doklady Biol Sci. 2006; 409: 346–348.
- Gao F, Gao Q, Duan X, Yue G, Yang A, et al. Cloning of an H+-PPase gene from Thellungiella halophila and its heterologous expression to improve tobacco salt tolerance. J Exp Bot. 2006; 57: 3259–3270. PubMed: https://pubmed.ncbi.nlm.nih.gov/16940040/
- Li X, Guo C, Gu J, Duan W, Zhao M, et al. Overexpression of VP, a vacuolar H+-pyrophosphatase gene in wheat (Triticum aestivum L.), improves tobacco plant growth under Pi and N deprivation, high salinity, and drought. J Exp Bot. 2014; 65: 683–696. PubMed: https://pubmed.ncbi.nlm.nih.gov/24474810/
- He C, Yan J, Shen G, Fu L, Holaday AS, et al. Expression of an Arabidopsis vacuolar sodium/proton antiporter gene in cotton improves photosynthetic performance under salt conditions and increases fiber yield in the field. Plant Cell Physiol. 2005; 46: 1848–1854. PubMed: https://pubmed.ncbi.nlm.nih.gov/16179357/
- Zhao FY, Zhang XJ, Li PH, Zhao YX, Zhang H. Co-expression of the Suaeda salsa SsNHX1 and Arabidopsis AVP1 confer greater salt tolerance to transgenic rice than the single SsNHX1. Mol Breed. 2006; 17: 341–353.
- Bhaskaran S, Savithramma DL. Co-expression of Pennisetum glaucum vacuolar Na+/H+ antiporter and Arabidopsis H+-pyrophosphatase enhances salt tolerance in transgenic tomato. J Exp Bot. 2011; 62: 5561–5570. PubMed: https://pubmed.ncbi.nlm.nih.gov/21841179/
- Peleg Z, Apse MP, Blumwald E. Engineering salinity and water stress tolerance in crop plants: getting closer to the field. Adv Bot Res. 2011; 57: 405–443.
- Rodríguez-Rosales MP, Gálvez FJ, Huertas R, Aranda MN, Baghour M, et al. Plant NHX cation/proton antiporters. Plant Signal. Behav. 2009; 4: 265–276. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2664485/
- Oh DH, Gong Q, Ulanov A, Zhang Q, et al. Sodium stress in the halophyte Thellungiella halophila, and transcriptional changes in a thsos1-RNA iterference line. J Integr Plant Biol. 2007; 49: 1484–1496.
- Pardo JM, Cubero B, Leidi EO, Quintero FJ. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. J Exp Bot. 2006; 57: 1181–1199. PubMed: https://pubmed.ncbi.nlm.nih.gov/16513813/
- Olías R, Eljakaoui Z, Li J, De Morales PA, Marín-Manzano MC, et al. The plasma membrane Na+/H+ antiporter SOS1 is essential for salt tolerance in tomato and affects the partitioning of Na+ between plant organs. Plant Cell Environ. 2009; 32: 904–916. PubMed: https://pubmed.ncbi.nlm.nih.gov/19302170/
- Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell 1996; 8: 617–627. PubMed: https://pubmed.ncbi.nlm.nih.gov/12239394/
- Shi H, Lee BH, Wu SJ, Zhu JK. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat Biotechnol. 2003; 21: 81–85. PubMed: https://pubmed.ncbi.nlm.nih.gov/12469134/
- Feki K, Quintero FJ, Khoudi H, Leidi EO, Masmoudi K, et al. A constitutively active form of a durum wheat Na+/H+ antiporter SOS1 confers high salt tolerance to transgenic Arabidopsis. Plant Cell Rep. 2014; 33: 277–288. PubMed: https://pubmed.ncbi.nlm.nih.gov/24150094/
- De Boer AH, Volkov V. Logistics of water and salt transport through the plant: structure and functioning of the xylem. Plant Cell Environ. 2003; 26: 87-101.
- Guo KM, Babourina O, Rengel Z. Na+/H+ antiporter activity of the SOS1 gene: life time imaging analysis and electrophysiological studies on Arabidopsis seedlings. Physiol Plant. 2009; 137: 155–165. PubMed: https://pubmed.ncbi.nlm.nih.gov/19758408/
- Mäser P, Eckelman B, Vaidyanathan R, Horie T, Fairbairn DJ, et al. Altered shoot/root Na+ distribution and bifurcating salt sensitivity in Arabidopsis by genetic disruption of the Na+ transporter AtHKT1. FEBS Lett. 2002; 531: 157–161. PubMed: https://pubmed.ncbi.nlm.nih.gov/12417304/
- Møller IS, Gilliham M, Jha D, Mayo GM, Roy SJ, et al. Shoot Na+ exclusion and increased salinity tolerance engineered by cell type- specific alteration of Na+ transport in Arabidopsis. Plant Cell. 2009; 21: 2163–2178. PubMed: https://pubmed.ncbi.nlm.nih.gov/19584143/
- Plett D, Safwat G, Gilliham M, Møller IS, Roy S, et al. Improved salinity tolerance of rice through cell type-specific expression of AtHKT1;1. PLoS ONE 2010; 5: e12571. PubMed: https://pubmed.ncbi.nlm.nih.gov/20838445/
- Mian A, Oomen RJ, Isayenkov S, Sentenac H, Maathuis FJ, et al. Over-expression of an Na+- and K+- permeable HKT transporter in barley improves salt tolerance. Plant J. 2011; 68: 468–479. PubMed: https://pubmed.ncbi.nlm.nih.gov/21749504/
- Horie T, Hauser F, Schroeder JI. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009; 14: 660–668. PubMed: https://pubmed.ncbi.nlm.nih.gov/19783197/
- Almeida P, Katschnig D, deBoer AH. HKT transporters-state of the art. Int J Mol Sci. 2013; 14: 20359–20385. PubMed: https://pubmed.ncbi.nlm.nih.gov/24129173/
- Maathuis FJM, Ahmad I, Patishtan J. Regulation of Na+ fluxes in plants. Front Plant Sci. 2014; 5: 467. PubMed: https://pubmed.ncbi.nlm.nih.gov/25278946/
- McAllister CH, Beatty PH, Good AG. Engineering nitrogen use efficient crop plants: the current status. Plant Biotechnol J. 2012; 10: 1011–1025. PubMed: https://pubmed.ncbi.nlm.nih.gov/22607381/
- Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012; 17: 458–467. PubMed: https://pubmed.ncbi.nlm.nih.gov/22658680/
- Kiba T, Feria-Bourrellier AB, Lafouge F, Lezhneva L, Boutet-Mercey S, et al. The Arabidopsis nitrate transporter NRT2.4 plays a double role in roots and shoots of nitrogen-starved plants. Plant Cell. 2012; 24: 245–258. PubMed: https://pubmed.ncbi.nlm.nih.gov/22227893/
- Little DY, Rao H, Oliva S, Daniel-Vedele F, Krapp A, et al. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc Natl Acad Sci. USA. 2005; 102: 13693–13698. PubMed: https://pubmed.ncbi.nlm.nih.gov/16157886/
- Krouk G, Lacombe B, Bielach A, Perrine-Walker F, Malinska K, et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev Cell. 2010; 18: 927–937. PubMed: https://pubmed.ncbi.nlm.nih.gov/20627075/
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, et al. Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci. U.S.A. 2011; 108: 18524–18529. PubMed: https://pubmed.ncbi.nlm.nih.gov/22025711/