More Information
Submitted: January 11, 2021 | Approved: February 01, 2021 | Published: February 02, 2021
How to cite this article: Toth M, Szabo Z, Toth Z. Alternative method for the transformation of Capsicum species. J Plant Sci Phytopathol. 2021; 5: 001-003.
DOI: 10.29328/journal.jpsp.1001053
ORCiD ID: orcid.org/0000-0002-3212-2438
Copyright License: © 2021 Toth M, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Alternative method for the transformation of Capsicum species
Mate Toth, Zoltan Szabo and Zoltan Toth*
Department for Genetics, Plant Genomics and Plant-Microbe Interaction Group, Agricultural Biotechnology Centre, NARIC, Hungary
*Address for Correspondence: Zoltan Toth, Agricultural Biotechnology Centre, NARIC, Szent-Györgyi Albert str. 4, Gödöllő, Hungary, H-2100, Tel: +36 (28) 526-100; Email: [email protected]
Capsicum (pepper) species have high economic values as vegetable crops and medicinal plants. Most of the Capsicum is known to be recalcitrant to plant regeneration in vitro, and to genetic transformation with Agrobacterium tumefaciens. However, genetic improvement against pathogens requires discovering new pest resistance genes and revealing their functions and mechanism in vitro. The development of improved transformation methods serves this purpose, which needs a binary vector technology carrying the gene of interest to be transferred into the host plants. Agrobacterium rhizogenes mediated transformation serves as a useful alternative way for the Capsicum transformation. The A. rhizogenes transformation compared to the A. tumefaciens transformation has the advantage that the method needs no regeneration step in vitro.
Our goal is to obtain a highly efficient transformation system that can be used to study the functions of different genes in Capsicum annuum varieties. Our study’s further goal is to validate and describe the candidate gene (Me1) involved in resistance against root-knot nematode species.
Capsicum species have a high economic value and have been serving as an important food crop worldwide for a long time. It is the second most significant crop in the family of Solanaceae after tomato [1]. The studying of pepper genes is having major barriers due to most of the Capsicum known to be recalcitrant to plant regeneration in vitro, and to genetic transformation with Agrobacterium tumefaciens [2]. A. tumefaciens mediated transformation in Capsicum species relies on the plant variety and showing low transformational efficiencies. Current studies indicate that finding A. tumefaciens mediated transformational protocols that can be applied across a wide range of Capsicum genotypes seems unrealistic at this moment [3,4].
A. rhizogenes (Gram-negative soil bacterium) carries a root-inducing plasmid including the Ri T-DNA (21,595 bp; EF433766) which cause the "hairy root" phenotype in plants [5,6]. The method has been extensively used also to produce secondary metabolites to study the functions of genes involved in root nodulation and root development, and confirm the functions of genes taking part in the resistance against root pathogens [7]. Concerning root diseases, root transformation with A. rhizogenes is a smart alternative to validate the function of candidate genes.
Root-knot nematodes, Meloidogyne spp. are cosmopolitan plant root parasitic nematodes. Nutrient losses and damages inflicted by the nematodes reduce root and shoot growth, photosynthetic capacity, and serious yield loss of infected plants [8]. More than 98 Meloidogyne species have been identified [9] and three of them (M. incognita, M. javanica, M. arenaria) are widely distributed and have broad ranges of host plants. During infection, the second-stage nematode juveniles (J2) invade the roots in the elongation zone and then migrate intercellularly and sedentarise into the differentiation zone of the plant vascular cylinder. In which, responding to stimulation from the parasite, few root cells located around the head of the nematode develop into specific feeding structures known as giant cells [10].
With the rapid change of climatic circumstances, there is a high risk of new pathogens will appear in our cultivated plants, so validating the functions of new resistance genes, and improving our cultivated plant's genetic basis has outstanding importance [11]. Plant resistance is the most efficient method of controlling root-knot nematodes, especially in Solanaceous crops including tomato (Solanum lycopersicum) and pepper (Capsicum annuum). Major resistance genes against RKN (root-knot nematodes) are available [8], and their application provides a safe and economically relevant strategy to control RKN. These resistance (R) genes generally harbor NBS–LRR (Nucleotide-Binding-Site Leucine-Rich Repeat) motifs and are often located in syntenic clusters in the genomes [12]. Introgression of resistance genes are the primary aim of pepper breeding programs because each of these genes provides resistance to the three major species of RKNs of M. incognita, M. arenaria and M. javanica.
Here we used a method developed for Radicles, hypocotyls, cotyledons of Capsicum annuum. Our final goal was to obtain a highly efficient A. rhizogenes transformation system for Capsicum annuum varieties. The further goal is to validate and describe the candidate gene (Me1) involved in resistance against root-knot nematodes of Capsicum annuum.
Plant materials
Pepper Capsicum annuum L. Global variety was applied. Surface sterilization of seeds: Capsicum seeds were surface sterilized with NaOCl (10%) for 15 mins followed by rinsing with double distilled and sterilized water.
Types of explants: Radicles, hypocotyls, cotyledons of the seedlings were scratched with a sterilized blade.
Inoculation: Agrobacterium rhizogenes ARqua1 were applied with coating the wound surface or with a thrust of a tungsten needle (1 µm tip) and cocultivated with the plants on hormone and antibiotics free Murashige & Skoog (MS) [13] media for 4 days then transferred to MPgy (MS base with 0,83 mg KJ, 50 mg NeFeEDTA, 100 mg inositol, 0,5 mg piridoxin, 0,5 mg thiamin, 0,5 mg NA, 0,5 mg IAA, 10 g glucose, 10 g sacharose, 10 g maltose with 6,5 g oxoid-agar) rooting medium with the respective selective antibiotics (Kanamycin).
Plasmid construction: For the transformation we used an empty gateway vector (pK7WG2D) which carried a Green fluorescent protein encoding reporter gene (GFP), to easily validate the transformant roots under fluorescent microscopy, and a kanamycin resistance gene.
Microscopy: Leica MZ10 F stereo microscopy with an external Leica EL6000 Light source for enhanced fluorescence imaging and visualized by the manufacturer’s own computer software.
In this study, we are trying to achieve an efficient way for validating genes involved in resistance against soil pathogens and genes which take part in the root nodulation and formation. Our research was based on the work of Aarrouf, et al. 2012, with modifications in the plant variety and inoculation method [14]. The development of “hairy root” phenotype (Figure 1) in A. rhizogenes mediated Capsicum transformants showed a thin filamentous structure (similar to the mycelia in fungi) especially along the surface of the cutting and pricked sites, however, not all transformant showed hairy root phenotype.
Figure 1: “Hairy-root” phenotype in Capsicum species.
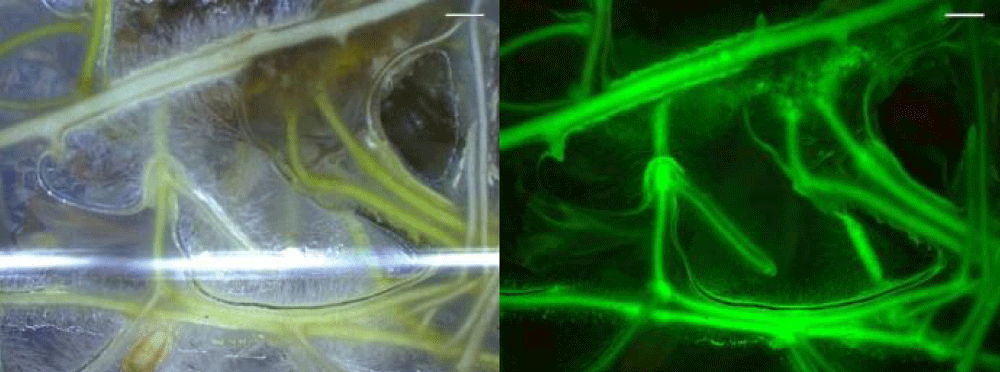
The other technical trouble was that non-transformant roots were also grown along with the taproot. In this case, removing them under a microscope can help to enhance the efficiency of the transformation. We successfully detected GFP signs in plant roots using fluorescent microscopy with both inoculation methods we used (Figure 2).
Figure 2: Expression of GFP in Capsicum annuum L. root tissues (Fluorescent microscopy image).
We concluded that the inoculated amount of bacteria turned out to be a crucial factor in the success of the method. Too many bacteria resulted in hard rooting and enhanced stem rotting while using too low amounts of bacteria resulted in a mosaic transformation (Figure 3) and weak gene expression.
Figure 3: Mosaic transformation in roots when using inadequate amount of inoculum.
The two different inoculation methods showed highly different efficiency. Coating the wound surface with bacteria showed a less effective transformation (lower number of transformed roots) and relatively low GFP expression but easy rooting of the inoculated explants. The thrust of a tungsten needle showed a high level of GFP expression with high transformation efficiencies however when we used too much bacteria for the infection it resulted in high plant material loss. Since the inoculation with the tungsten needle showed a higher transformation efficiency and expression rates, therefore, this method was preferred over coating the wound surface.
Different explants were transformed with different efficiencies. The cotyledon showed the highest transformational efficiency (70%), however, due to the rooting on the cotyledon, the plants became distorted and the plant material became difficult to handle. The hypocotyl transformed with high efficiency (60%), while in the case of the radicle it was less efficient (lower yield of transformant roots, low GFP expression) compared to the other two explants (50%). Our results suggest that hypocotyl transformation proved to be the most effective method, which coincides with the results of Aarrouf, et al. 2012.
The advantages of this method are high efficiency and relatively fast transformation. In conclusion, we have developed a highly efficient alternative method for A. rhizogenes-mediated transformation of Capsicum annuum L. Global variety which showed a higher transformation efficiency, than other transformation methods.
The best co-transformation efficiency was obtained for hypocotyl explants using a tungsten needle for the infection. However it is really hard to measure the right amount of bacteria for the transformation with the tungsten needle, we found out that if the tungsten needle tip was gently pinched to a 1-day old bacterial lawn the transformation was efficient and we could yield a high amount of plants with transformed roots.
Hairy root culture has significant advantages such as rapid growth and reproducibility, and sustained maintenance of co-transformed roots is possible by vegetative propagation. This approach based on A. rhizogenes will support an intensive study of host–pathogen interactions and functional analysis of potential resistance genes. In the future, we would also like to validate if our system can be applied for a wide range of Capsicum genotypes.
The authors wish to thank Prof. G Gyulai for his comments on the manuscript. This work was supported by the ÚNKP-19-II-3 of the New National Excellence Program of the Ministry of Human Capacities.
- Hasan MJ, Kulsum MU, Ullah MZ, Hossain MM, Mahmud ME. Genetic diversity of some chili (Capsicum annuum L.) genotypes. Int J Agric Res Innovat Technol. 2014; 4: 32-35.
- Wolf D, Matzevitch T, Steinitz B, Zelcer A. Why is it difficult to obtain transgenic pepper plants? Acta Horticulturae. 2001; 560: 229-233.
- Li D, Zhao K, Xie B, Zhang B, Luo K. Establishment of a highly efficient transformation system for pepper (Capsicum annuum L.). Plant Cell Rep. 2003; 21: 785-788.
- Steinitz B, Wolf D, Matzevitch-Josef T, Zelcer A. Regeneration in vitro and genetic transformation of pepper (Capsicum spp.): The current state of art. Capsicum and Eggplant Newsletter.1999; 18: 9-15.
- Setamam N Md, Sidik NJ, Rahman ZA, Zai CRCM. Induction of hairy roots by various strains of Agrobacterium rhizogenes in different types of Capsicum species explants. BMC Res Notes. 2014; 7: 1–8. PubMed: https://pubmed.ncbi.nlm.nih.gov/24981787/
- Desmet S, Dhooghe E, De Keyser E, Quataert P, Eeckhaut T, et al. Segregation of rol genes in two generations of Sinningia speciosa engineered through wild type Rhizobium rhizogenes. Front Plant Sci. 2020; 11: 859. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7333734/
- De Paolis A, Frugis G, Giannino D, Iannelli MA, Mele G, et al. Cellular and molecular biotechnology: Following Mariotti’s steps. Plants (Basel). 2019; 8: 18. PubMed: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6359066/
- Trudgill DL. Resistance to and tolerance of plant parasitic nematodes in plants. Annual Review of Phytopathology. 1991; 29: 167–l 93.
- Jones JT, Haegeman A, Danchın EGJ, Gaur HS, Helder J, et al. Top 10 plant parasitic nematodes in molecular plant pathology. Mol Plant Pathol. 2013; 14: 946–961 PubMed: https://pubmed.ncbi.nlm.nih.gov/23809086/
- Bird AF. The ultrastructure and histochemistry of a nematode-induced giant-cell. J Biophys Biochem Cytol. 1961; 11: 701–715. PubMed: https://pubmed.ncbi.nlm.nih.gov/13869341/
- Reddy KR, Hodges HF. (Eds.). Climate change and global crop productivity. Biologia Plantarum. 2000; 43. CABI.
- Djian-Caporalino C, Fazari A, Arguel MJ, Vernie T, Casteele CV, et al. Root-knot nematode (Meloidogyne spp.) Me resistance genes in pepper (Capsicum annuum L.) are clustered on the P9 chromosome. Theor Appl Genet. 2007; 114: 473–486.
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962; 15: 473–449.
- Aarrouf J, Castro-Quezada P, Mallard S, Caromel B, Lizzi Y, et al. Agrobacterium rhizogenes-dependent production of transformed roots from foliar explants of pepper (Capsicum annuum): a new and efficient tool for functional analysis of genes. Plant Cell Rep. 2012; 31: 391-401. PubMed: https://pubmed.ncbi.nlm.nih.gov/22016085/