Table of Contents
Primer Pairs for Rice (Oryza sativa L.) Bisulfite Sequencing Studies
Published on: 6th November, 2018
OCLC Number/Unique Identifier: 7905997684
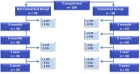
Many agriculturally important properties such as heterosis, inbreeding depression, phenotypic plasticity, and resistance for biotic and abiotic stresses are thought to be affected with epigenetic components. New discoveries related with epigenetics are likely to have a major impact on strategies for crop improvement in rice breeding. However, assessing the contribution of epigenetics to heritable variation in plant species still poses major challenges. Methylation of cytosine in DNA is one of the most important epigenetic mechanisms in plants. DNA methylation not only plays significant roles in the regulation of gene activity, but also it is related with genomic integrity. Although most of next generation sequencing (NGS) technologies do not require the use of target specific primer pairs to identify and study DNA cytosine methylation, validation studies of NGS uses selective primer pairs. Bisulfite sequencing technique is a gold method for DNA cytosine methylation studies. However, bisulfite sequencing requires the development of bisulfite primer pairs to selectively study DNA sequences of interest. In this study 9 bisulfite specific primer pairs were identified and validated. These primer pairs successfully amplified bisulfite converted and unconverted genomic DNA extracted from radicle and plumule of rice (Oryza sativa L.) seedlings. Results of the present study clearly revealed the occurrence of CG, CHG and CHH (H stands for C. T or A nucleotides) contents in studied DNA sequence targets were different indicating potential role of DNA cytosine methylation in these genes. Primer pairs reported in this study could be used to detect DNA methylation which is one of the most important epigenetic mechanisms affecting the development, differentiation or the response to biotic and abiotic stress in rice (Oryza sativa L.).
Use of essential oils as new food preservatives (Case: Eucalyptus grandis and Eucalyptus crebra)
Published on: 2nd November, 2018
OCLC Number/Unique Identifier: 7912408220
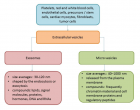
This study purposed to study the Preservative agents that are required to ensure that manufactured foods remain safe and unspoiled; work was conducted to evaluate the efficacy of essential oils from two eucalyptus species, Eucalyptus grandis and Eucalyptus crebra in food preservatives; to run this experiment flesh eucalyptus leaves collected from Ruhande Arboretum forest were submitted to hydrodistillation and yields(amount) of 0.38 and 0.34 % for Eucalyptus grandis and Eucalyptus crebra were obtained, respectively. Phaseolus vulgaris, Sorghum condatum, cooked Ipomoea batatas (sweet potatoes) and bread were the sample foods used to assess their preservative efficacy. Acanthoscelides obtectus and Stophilus oryzae were used as pests for Phaseolus vulgaris and Sorghum condatum respectively. For bread and cooked Ipomoea batatas, Rhizopus nigricans are used to assess the efficacy of these two essential oils to inhibit their growth; the obtained results revealed that those essential oils could act as insecticide in the storage of Phaseolus vulgaris and Sorghum condatum. Essential oil from Eucalyptus grandis protected these two foods against pests in the periods of 4 and 9 days, respectively while essential oil from Eucalyptus crebra protected them for the periods of 6 and 11days, respectively.
Advances in research of the structural gene characteristics of the aflatoxin biosynthetic gene cluster
Published on: 31st October, 2018
OCLC Number/Unique Identifier: 7912359098
Aflatoxins, produced by Aspergillus spp., are strongly toxic and carcinogenic fungal secondary metabolites. Aflatoxin biosynthesis is a complex process and involves at least 30 genes clustered within an approximately 75 kB gene cluster. In this paper, we reviewed current status of the researches on the characterized structural genes involved in aflatoxin biosynthesis and their roles in aflatoxin-producing fungi, especially in A. flavus and A. parasiticus, which will improve our understanding of the mechanism of aflatoxin biosynthesis and regulation and provide reference for further study.
Bacillus amyloliquefaciens as a plant growth promoting bacteria with the interaction with of grass salt Distichlis palmeri (Vasey) under field conditions, in desert of Sonora, Mexico
Published on: 2nd October, 2018
OCLC Number/Unique Identifier: 7900050235
The halophyte Distichlis palmeri (Vasey) is a plant resource with high potential to be harvested in the coastal areas of northwestern Mexico; enlarge the knowledge and domestication for its incursion into the agricultural sector, plays an important role for arid areas with saline intrusion problems. However, its productivity depends on the supplementary supply of nitrogen, as well as other essential macro and micronutrients. The microorganisms considered beneficial are an alternative to chemical fertilization, highlighting those Plant Growth Promoting Bacteria (PGPB). In the present study, the inoculation of the Bacillus amyloliquefaciens (B.a.) as a halobacterium PGPB was evaluated to know the response in seeds of Distichlis spicatai obtained from natural population from colorado river in Delta north of the Gulf of California. Wild seed was collected and germinated previously inoculated with B. a., and sowed in germinated beds. Later, seedlings were planted under field and salinity conditions in the coast of Hermosillo, Sonora. Three treatments were examined (T1: B.a., T2: Chemical fertilization, T3: Negative control), with four repetitions each treatment. Each repetition consisted of experimental plots of 5 x 5 m, with a separation of 1 m between them. The harvest was carried out 600 days after sowing. The results indicate that treatments inoculated with halobacteria B.a., showed significant results in crude protein, non-protein nitrogen, neutral detergent fiber and acid detergent fiber, as well as spike length and number of seeds. The results obtained suggests the feasibility of biofertilizers where biomass and seed production are significant compared to non-inoculated controls.

HSPI: We're glad you're here. Please click "create a new Query" if you are a new visitor to our website and need further information from us.
If you are already a member of our network and need to keep track of any developments regarding a question you have already submitted, click "take me to my Query."