More Information
Submitted: March 19, 2024 | Approved: April 12, 2024 | Published: April 15, 2024
How to cite this article: Vargas LMT, Pérez AH, Valencia ASH. New Fungi Associated with Blackberry Root Rot (Rubus spp.), in Michoacán, Mexico. J Plant Sci Phytopathol. 2024; 8: 038-040.
DOI: 10.29328/journal.jpsp.1001129
Copyright License: © 2024 Vargas LMT, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Necrosis; Wilting; Fusarium spp.; Agricultural management
New Fungi Associated with Blackberry Root Rot (Rubus spp.), in Michoacán, Mexico
Luis Mario Tapias Vargas1, Anselmo Hernández Pérez1 and Adelaida Stephany Hernández Valencia2*
1Uruapan-INIFAP Experimental Field, Latin American Avenue 1101, Uruapan, Michoacán 60150, Mexico
2Postgraduate in Phytopathology, Postgraduate College, Montecillo Campus, Km. 36.5 Carretera México-Texcoco, C.P. 56230, Texcoco, State of Mexico, Mexico
*Address for Correspondence: Adelaida Stephany Hernández Valencia, Postgraduate in Phytopathology, Postgraduate College, Montecillo Campus, Km. 36.5 Carretera México-Texcoco, C.P. 56230, Texcoco, State of Mexico, Mexico, Email: [email protected]
Los Reyes, Michoacán, Mexico, is one of the main blackberry-producing places in the world, however, the disease located at the root level has caused important economic losses. Currently has been reported that the fungus Fusarium spp., is the main causal agent but actions to control it have failed. The objective of this work was to identify the possible presence of unreported pathogenic fungi in the root system of the blackberry and identify them molecularly. It was sampled in a commercial open-air orchard from Los Reyes, pieces of roots were taken from symptomatic plants with wilting and decay. The fungi were isolated in the laboratory, identified with taxonomic keys, extraction was performed, and the sequences obtained were compared with those reported in the NCBI gene bank. Among the results obtained were Kalmusia italica, Epicoccum nigrum, Microsphaeropsis arundinis, Achizophyllum commune, and, as expected, some species of Fusarium spp.
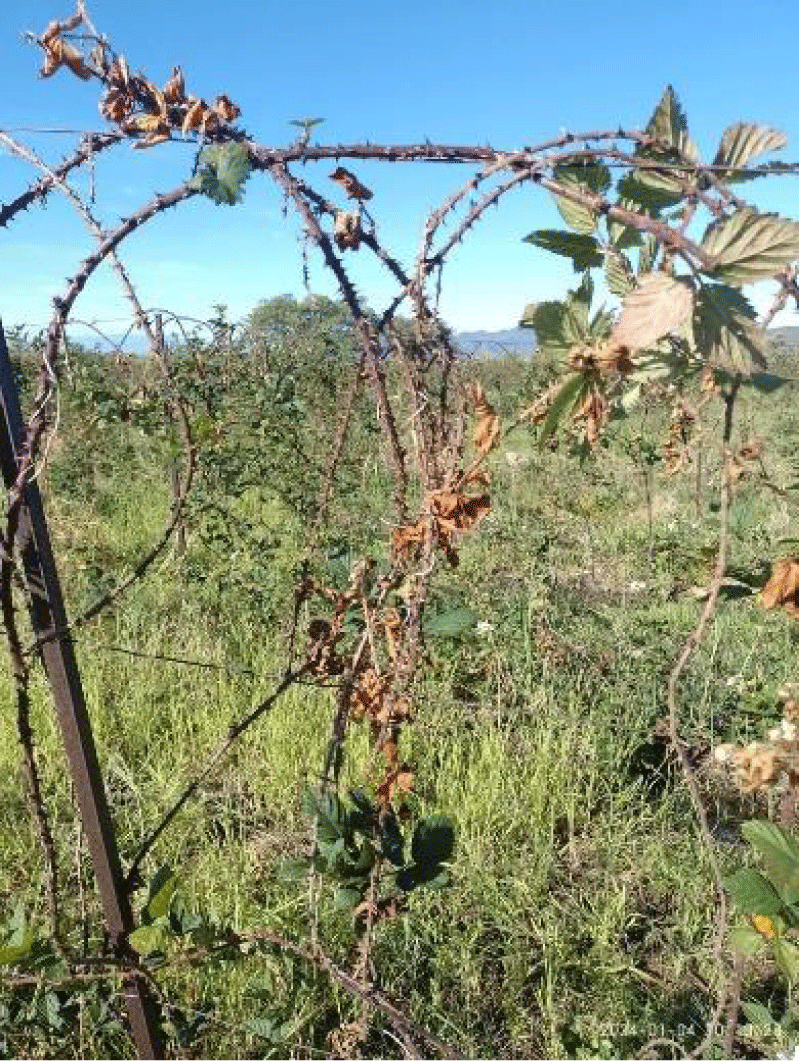
Mexico is the main blackberry-producing country worldwide with a production of over 215,923 tons, where the state of Michoacán contributes 97% of this berry fruit [1]. Among the main diseases that have been reported for this crop are gray mold (Botrytis cinerea), orange rust (Arthuriomyces peckianus), leaf spot (Cercospora sp), anthracnose (Colletotrichum gloesporoides), mildew (Peronospora sparsa), and root rot (Fusarium sp) [2]. The genus Fusarium damages the root system of the crop and has caused land abandonment, affecting more than 2,000 hectares. Various investigations have reported other fungi associated with the symptoms of root and crown rot of the blackberry plant, such as Rhizoctonia sp and Phytophthora spp. (Figure 1). However, the management of these fungi with specific fungicides has not affected preventing the death of the plant, so other unidentified fungi may be involved. The objective of this work was to identify the possible presence of unreported pathogenic fungi in the root system of the blackberry and identify them molecularly, from the state of Michoacán.
Figure 1: Death of the foliage of the blackberry plant (Rubus spp), due to root rot.
Samples of blackberry plant roots with visible symptoms of wilting were collected at a depth of 30 cm at equidistant points. The root was fragmented into pieces no larger than 0.5 cm, including healthy and diseased tissue. They were disinfected with 3.0% sodium hypochlorite and were seeded in Petri dishes at 28 °C in total darkness. Subsequently, purification and multiplication were conducted by the hyphal tip in triplicate. Death foliage from the pathogens obtained was identified morphologically according to the keys of Lévesque & Cock, [3]; Leslie and Summerell [4] and molecularly using the PCR-ITS technique, extracting DNA according to the methodology of Doyle and Doyle [5], from 0.2 g of mycelium from the pure culture with lysis buffer (EDTA 50 mM, pH 8.5; 100 mM Tris HCl, pH 8; 50 mM NaCl; 2% SDS). DNA visualization was performed on 2.0% agarose gel stained with GelRed (GenScript®). The amplification of the ITS region was conducted with different ITS primers. Likewise, the reaction product will be visualized employing electrophoresis in a 2.0% agarose gel stained with GelRed (GenScript®) and the PCR product was sequenced at the Phytopathology Laboratory of the Universidad Autonoma Agraria Antonio Narro, with the dideoxynucleotide method labeled on the model 3500 and 3130, Genetic Analyzer sequencers (Applied Biosystems), and subsequently obtain the molecular identification. Isolation of fungal DNA and its Polymerase Chain Reaction (PCR) was the basic step to identify 18S rRNA. DNA was isolated, its purity was checked using NANODROP, it was amplified by PCR and the 18S rRNA was sequenced. To identify the fungi isolated from the blackberry root, the partial sequence of the 18S rRNA gene was compared with the complete sequence available in the GenBank database using a BLAST search (NCBI).
The results indicated that fungi previously reported in other investigations associated with the symptom of root rot in blackberries were found, which were Fusarium oxysporum and Fusarium subglitinans. This study, using the PCR technique and genetic sequencing, allowed us to discover new, unreported pathogenic fungi associated with said symptoms and plant death, such as the fungi Kalmusia italica, Epicoccum nigrum, Microsphaeropsis arundinis and Achizophyllum commune. The discovery of these new, unreported fungi (Table 1) makes it possible to develop control methodologies to determine the affected crop area and prevent or recover planting in soils with these pathogens. Among the results obtained, fungi previously found in other investigations associated with the symptom of root rot in blackberries were found, which were Fusarium oxysporum, and Fusarium subglitinans. However, an interesting result is finding other new fungi associated with said symptoms, such as fungi Kalmusia italica, Epicoccum nigrum, Microsphaeropsis arundinis, and Schizophyllum commune.
| Table 1: Molecular characterization of the isolates from the sequences reported in the gene bank with the Intergenic Sequences (ITS) of the rDNA genes. | |||
| Isolate1 | Fungi | Accesion2 | Similarity (%)3 |
| A1-1_E03 | Kalmusia italica | KY702577 | 98.9 |
| A1-4_F03 | Kalmusia italica | KY702577 | 91.8 |
| A10-1_G05 | Fusarium oxysporum | HQ451892 | 99.6 |
| A10-4_H05 | Fusarium oxysporum f. sp. ciceris | DQ906172 | 76.5 |
| A11-1_A06 | Fusarium oxysporum | KJ584545 | 99.6 |
| A3-4_B04 | Epicoccum nigrum | MT441593 | 92.0 |
| A4-1_C04 | Microsphaeropsis arundinis | MW760784 | 94.2 |
| A4-4_D04 | Microdiplodia sp. | KY977605 | 96.2 |
| A5-1_E04 | Fusarium oxysporum | MN559971 | 99.6 |
| A6-1_G04 | Schizophyllum commune | ON329677 | 97.8 |
| A6-4_H04 | Schizophyllum commune | ON329677 | 94.2 |
| A7-1_A05 | Fusarium oxysporum | KM203588 | 99.4 |
| A7-4_B05 | Fusarium subglutinans | KY318486 | 95.6 |
| 1Nomenclature for the different isolates.2Accession number in the NCBI (National Center of Biotechnology Information) database. 3Similarity index between the sequences of the isolated species and the compare. | |||
Kalmusia italica presents pycnidia solitary, gregarious or grouped, superficial on PDA, and ostiolate. Pycnidial wall up to 50 μm - 90 μm wide, comprising several layers of pseudoparenchymatous, cells of textura angularis and prismatica, the outer layer composed of thick-walled, dark brown cells, lighter towards the inner layers of hyaline cells. Conidiogenous cells 3.4 – 6.2 × 2 – 3.5 μm (x = 4.8 × 3.1 μm, n = 20), short cylindric, conidiogenous holoblastic, hyaline, smooth. Conidia 1 – 1.5 × 2 – 3.4 μm (x = 1.2 × 2.9 μm, n = 25), oblong, ellipsoid-cylindric, aseptate, hyaline to lightly pigmented, smooth-walled [6]. Epicoccum nigrum contains colonies that are fast-growing, suede-like to downy, with a strong yellow to orange-brown diffusible pigment. When sporulating, numerous black sporodochia (aggregates of conidiophores) are visible. Conidia are formed singly on densely compacted, non-specialized, determinant, slightly pigmented conidiophores. Conidia are globose to pyriform, mostly 15 µm - 25 µm diameter with a funnel-shaped base and broad attachment scar, often seceding with a protuberant basal cell. Conidia become multicellular (dictyoconidia), darkly pigmented, and have a verrucose external surface [7]. M. arundinis is distinguished from the species M. olivacea and M. callista by its smooth, thin-walled, cylindrical, guttulate conidia measuring 4 to 4.5 by 1.5 m [8]. Schizophyllum commune is one of the most widely distributed fleshy fungi (teleomorphic state), often found on dead and decaying organic matter, especially rotten wood of trees. It belongs to the Phylum Basidiomycota, Subphylum Agaricomycotina, Order Agaricales which contains fungi, colloquially known as mushrooms. The distinctive feature of these fungi is the formation of a macroscopic fructification, the basidiocarp, that contains basidiospores (sexual spores) developing on the outside of a club-shaped or elongated structure called the basidium [9].
The isolated and identified fungus K. italica was screened for its tolerance toward heavy metals in a PDA medium containing heavy metals from 50 to 250 ppm. The fungi showed various resistant strategies toward the heavy metals of different concentrations. The result showed that the response of the isolates to heavy metal depends on the metal evaluated, its concentration in the medium, and the isolate. Some isolates were tolerant, while others reacted negatively even at low metal ion concentrations [10], however, we must evaluate the plant's tolerance to the fungus. As biological control, Epicoccum nigrum is a saprophytic or endophytic fungus that is found worldwide. Because of the antagonist effects of E. nigrum on many plant pathogens, current studies on E. nigrum have focused on the development of biological control agents and the utilization of its various metabolites [11]. Microsphaeropsis arundinis is related to the class of Coelomycetes fungi. According to the literature, this fungus is associated with rare infections of the skin [12]. Microsphaeropsis species are typically found as saprobes and parasites of terrestrial plants. They inhabit branches and leaves of various plant hosts and are ubiquitous in soil and freshwater environments [8]. S. commune has been reported to be a pathogen of humans and trees, but it adopts a saprobic lifestyle by causing white rot [13]. It is found on fallen branches and timber of deciduous trees. At least 150 genera of woody plants are substrates for S. commune, but it also colonizes softwood and grass silage [14].
Although this advanced research requires carrying out Koch's postulates (pathogenicity tests), to determine if in reality, these fungi found associated with the symptom can generate potential damage to blackberry roots, it was urgent to know if there were more fungi associated with the symptom. withering, which this work reliably verifies. The preliminary identification of phytopathogens based on taxonomic studies of fungi and their host pathology [15] requires considerable time and experience; however, molecular methods such as PCR speed up identification by analyzing specific regions within genes. The sequences of Internal Transcribed Spacers (ITS) and regions such as elongation factor 1-alpha (TEF 1-α) are the most used in identification, as a standard barcode for fungi, [16] and once identified, take measures to control their proliferation.
Various investigations have determined one species of Fusarium spp., as the main causal agent of root rot in blackberries, however, it is important to know those fungi that are associated with the disease, which can form a complex for the development of the symptoms can either function as biological control agents, or not generate pathogenesis in the plant. Molecular identification is a tool that accelerates the identification process and has a better command of the integrated management of the disease. The fungi Kalmusia italica, Epicoccum nigrum, Microsphaeropsis arundinis, and Achizophyllum commune, found in this research, must undergo pathogenicity tests, and determine their role in blackberry root rot in Michoacán.
- SADER. 2019. https://www.gob.mx/agricultura/colima/articulos/la-zarzamora-cada-dia-mas-mexicana-235435?idiom=es
- Fernández-Pavía SP, Rodríguez-Alvarado GR, Gómez-Dorantes N, Gregorio-Cipriano MR y Fernández-Pavía YL. Enfermedades en plantas en el estado de Michoacán. Biológicas. 2012; 14(2):75-89.
- Lévesque CA, Harlton CE, de Cock AW. Identification of some oomycetes by reverse dot blot hybridization. Phytopathology. 1998 Mar;88(3):213-22. doi: 10.1094/PHYTO.1998.88.3.213. PMID: 18944967.
- Leslie JF, Summerell BA. Fusarium laboratory workshops-A recent history. Mycotoxin Res. 2006 Jun;22(2):73-4. doi: 10.1007/BF02956766. PMID: 23605575.
- Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990; 12:13-15.
- Liu JK, Hyde KD, Jones EG, Ariyawansa HA, Bhat DJ, Boonmee S, Camporesi E. Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal diversity. 2015; 72:1-197.
- Ellis MB. Dematiaceous hyphomycetes. Commonwealth Mycological Institute, Kew. 1971; 608.
- Pendle S, Weeks K, Priest M, Gill A, Hudson B, Kotsiou G, Pritchard R. Phaeohyphomycotic soft tissue infections caused by the coelomycetous fungus Microsphaeropsis arundinis. J Clin Microbiol. 2004 Nov;42(11):5315-9. doi: 10.1128/JCM.42.11.5315-5319.2004. PMID: 15528731; PMCID: PMC525150.
- Chowdhary A, Randhawa HS, Gaur SN, Agarwal K, Kathuria S, Roy P, Klaassen CH, Meis JF. Schizophyllum commune as an emerging fungal pathogen: a review and report of two cases. Mycoses. 2013 Jan;56(1):1-10. doi: 10.1111/j.1439-0507.2012.02190.x. Epub 2012 Apr 23. PMID: 22524529.
- Sumathi S, Priyanka V, Krishnapriya V, Suganya K. Identification of a novel strain of fungus Kalmusia italica from untouched marine soil and its heavy metal tolerance activity. Bioremediation Journal. 2021; 25(2):91-107.
- Li T, Im J, Lee J. Genetic Diversity of Epicoccum nigrum and its Effects on Fusarium graminearum. Mycobiology. 2022 Dec 13;50(6):457-466. doi: 10.1080/12298093.2022.2148394. PMID: 36721792; PMCID: PMC9848293.
- Botero WB, Amorim MRD, Carlos IZ, Polesi MC, Santos LCD. Aromatic Polyketides and Macrolides from Microsphaeropsis arundinis. Journal of the Brazilian Chemical Society. 2020; 31:364-369.
- Schmidt O, Liese W. Variability of wood degrading enzymes of Schizophyllum commune. Holzforschung. 1980; 34:67–72.
- De Jong JF. Aerial Hyphae of Schizophyllum commune: Their Function and Formation. PhD thesis, Univ. Utrecht. 2006.
- Aoki T, O´Donnell K, Scandiani MM. Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. Nov., F. cuneirostrum sp. Nov., F. tucumaniae, and F. virguliforme. Mycoscience. 2005; 46:162-183.
- Fernández-Ortuño D, Loza-Reyes E, Atkins SL, Fraaije BA. The CYP51C gene, a reliable marker to resolve interspecific phylogenetic relationships within the Fusarium species complex and a novel target for species-specific PCR. Int J Food Microbiol. 2010 Dec 15;144(2):301-9. doi: 10.1016/j.ijfoodmicro.2010.10.013. Epub 2010 Oct 21. PMID: 21071105.