More Information
Submitted: May 26, 2023 | Approved: June 09, 2023 | Published: June 10, 2023
How to cite this article: Otun S, Achilonu I, Ntushelo K. The secondary metabolites profiling of the phytopathogenic fungus Sclerotinia Sclerotiorum. J Plant Sci Phytopathol. 2023; 7: 027-038.
DOI: 10.29328/journal.jpsp.1001102
Copyright License: © 2023 Otun S, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Sclerotinia sclerotiorum; Secondary metabolites; Metabolic pathways; Pathogenicity; Virulence factors
The secondary metabolites profiling of the phytopathogenic fungus Sclerotinia Sclerotiorum
Sarah Otun1* , Ikechukwu Achilonu1
, Ikechukwu Achilonu1 and Khayalethu Ntushelo2
and Khayalethu Ntushelo2
1School of Molecular and Cell Biology, Faculty of Science, Protein Structure-Function and Research Unit, University of the Witwatersrand, Braamfontein, Johannesburg, South Africa
2Department of Agriculture and Animal Health, Science Campus, University of South Africa, Florida, South Africa
*Address for Correspondence: Sarah Otun, School of Molecular and Cell Biology, Faculty of Science, Protein Structure-Function and Research Unit, University of the Witwatersrand, Braamfontein, Johannesburg, South Africa, Email: [email protected]
Sclerotinia sclerotiorum is a necrotrophic plant pathogen causing more than 60 different disease symptoms in approximately 400 plants globally. Hence, due to this distinctive characteristic, S. sclerotiorum has been the subject of various research to comprehend its pathogenicity mechanism, including virulent genes, proteins, and metabolites. Likewise, the genomic annotation of S. sclerotiorum uncovered its remarkable potential for producing secondary metabolites, of which genome mining has additionally prompted the disclosure of these uncharacterized metabolic pathways, which might aid the pathogenicity process. To comprehend the secondary metabolites secreted by S. sclerotiorum that might be involved in its pathogenicity, a secondary metabolite-level investigation of this plant pathogen was performed. Profiling and characterizing these secondary metabolites produced during in vitro germination would increase the current knowledge of this pathogen.
In this study, S. sclerotiorum secondary metabolites profile examination was conducted, utilizing the Ultra-High Resolution Qq-Time-Of-Flight mass spectrometer (UHR-QqTOF). Proficient data analysis and verification with the genomic pathways of S. sclerotiorum gave an unequivocal metabolome profile of this pathogen. Two hundred and thirty secondary metabolites were identified in all three biological replicates, and their bodily functions were identified.
Sclerotinia sclerotiorum is a necrotrophic plant pathogen that is the causative agent of approximately 60 symptomatic diseases, including the notorious Sclerotinia stem rot, drop, crown rot, blossom blight, and white mould (which is the most prevalent) [1,2].
Sclerotinia sclerotiorum is a highly destructive phytopathogenic fungus that poses a significant threat to numerous plant species [3]. This fungus is responsible for causing Sclerotinia stem rot or white mould, a disease that results in considerable yield losses and economic damage in agriculture. The pathogenic mechanisms employed by S. sclerotiorum involve the production of enzymes and toxins [4]. It secretes various cell wall-degrading enzymes, such as polygalacturonases and cellulases, which break down plant cell walls, facilitating fungal penetration and colonization [5]. Oxalic acid, another virulence factor the fungus produces, contributes to tissue maceration and cell death [6]. These mechanisms collectively lead to symptoms including wilting, stem cankers, water-soaked lesions, and the formation of characteristic white cottony mycelium and sclerotia on infected tissues [6,7].
To combat S. sclerotiorum, a range of strategies for prevention and control are currently employed. Cultural practices play a vital role, including crop rotation with non-host plants to disrupt the pathogen's life cycle and reduce inoculum levels in the soil. Proper spacing between plants improves air circulation and reduces humidity, creating an unfavorable environment for fungal growth. Timely and appropriate irrigation practices minimize plant wetness and limit disease development [4]. Chemical control utilizing fungicides, such as boscalid, iprodione, and thiophanate-methyl, has demonstrated efficacy in managing the disease [8]. However, adopting integrated pest management practices and considering potential environmental impacts is essential [4,9].
Biological control agents, including certain species of Trichoderma and Bacillus, show promise in suppressing S. sclerotiorum [10]. These beneficial microorganisms compete with the pathogen for resources, produce antimicrobial compounds, and induce plant defense mechanisms. Furthermore, breeding programs for resistant cultivars offer a long-term and sustainable solution. Breeders aim to reduce crop susceptibility to the disease by incorporating resistance traits into commercial varieties. However, the complex nature of host-pathogen interactions presents challenges in achieving broad-spectrum resistance [11].
Hence, a comprehensive approach combining cultural practices, chemical control, biological agents, and host resistance is essential for effectively managing S. sclerotiorum.
This epidemic has opened S. sclerotiorum to broad research, including its biology, genomic analysis, and proteome-level studies [6,7,12]. All these are proposed for systematic searches for its molecular characteristics and the bases of its pathogenicity [13,14]. More needs to be done to investigate the metabolites secreted by this necrotrophic pathogen. Consequently, the collection of metabolites involved in the necrotrophic lifestyle of this pathogen remains vague, thereby creating a gap in the available knowledge on this pathogen.
Given these perceptions, it could be hypothesized that S. sclerotiorum produces numerous metabolites that could be by-products of the proteins. Henceforth, this investigation aims to profile secondary metabolites produced during in vitro germination of S. sclerotiorum, characterize them based on their functions, and discover the pathways in which they are implicated. The discovery of such metabolites would fill the knowledge vacuum of S. sclerotiorum metabolomics, creating opportunities for novel metabolite disclosures.
Metabolites were extracted from the actively growing mycelia of S. sclerotiorum as described by [15]. They were profiled utilizing the automated Ultra-High Resolution Qq-Time-Of-Flight mass spectrometer that generated chromatograms from detected chemical compounds available in the extract. This step was then preceded by data analysis using the Magma web tool to interpret and identify the secondary metabolites from the chromatogram.
Finally, the identified metabolites were classified based on their chemical composition and biological functions.
Fungi sample preparation and metabolite extraction
The virulent S. sclerotiorum wild-type strain 1980 UF-70 acquired from the Agricultural Research Council – Plant Protection Research, Tshwane, was utilized for this investigation. Mycelia of the fungus were harvested from a 5-day-old culture growing on potato dextrose agar at temperatures ranging from 4 ºC to room temperature. Harvested mycelia were ground in liquid nitrogen using a mortar and pestle, followed by a metabolite extraction technique.
Extraction of metabolites secreted by S. sclerotiorum was performed according to [16], with slight modification. Fifty milligrams of the powdered S. sclerotiorum mycelia samples were weighed, and then 1.5 mL methanol: water (75%:25%, v/v) was added to the samples, following ultrasonic blending for 5 minutes. The mixtures were centrifuged at 12,000 rpm for 15 minutes at 4 ºC [17] and the supernatants were dispensed into 1.5 mL centrifuge tubes for subsequent mass spectrometry investigation.
Analysis of metabolites extracted from Sclerotinia sclerotiorum
One microliter of S. sclerotiorum metabolites extracts was separated using an RP C18 column (50 x 2 mm, 1.7 µm particle size) on UHR-QqTOF (Bruker Daltonics). The system was connected to a networked series printer for recording chromatograms, Chromeleon Data System (Thermoscientific). The following gradient was utilized for the separation; the flow rate was 400 μL/min using (A) water + 0.1% HCOOH (B) Acetonitrile + 0.1% HCOOH as the mobile phase. The gradient was at 0 minutes 1% B; 1 minute 1% B; 10 minutes 99% B; 12 minutes 99% B; 12.5 minutes 1% B; 14 minutes 1% B ESI-MS measurements were performed using positive ionization on the maXis UHR-QqTOF MS m/z range: 100- 1200 m/z, acquisition rate: 3, 5, 10, 20 Hz [18].
Data analysis was done utilizing the MAGMa web tool https://www.emetabolomics.org/ according to [19]. Chromatogram generated by the UHR-QqTOF was queried against the KEGG compound database and the PubChem database, respectively, excluding peaks corresponding to contaminants, solvents, or media used.
Lastly, the functional characterization of the identified secondary metabolites was done utilizing the MetaboAnalyst and KEGG BRITE resources [20].
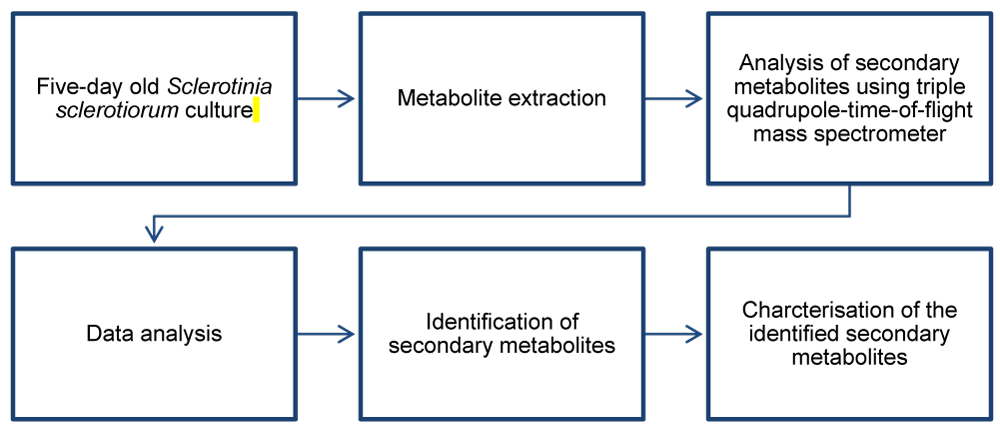
The workflow for characterizing the S. sclerotiorum metabolome is shown in Figure 1.
Figure 1: A schematic workflow for analyzing secondary metabolites produced by Sclerotinia sclerotiorum.
Pathway analysis of identified secondary metabolites produced by Sclerotinia sclerotiorum
All the secondary metabolites identified above were subjected to the pathway analysis module of the MetaboAnalyst web tool. MetaboAnalyst pathway analysis module utilizes the results from pathway enrichment (using hypergeometric test) and pathway topology analysis to detect the most significant pathways in the present investigation [21].
In the present investigation, 230 secondary metabolites were discovered in the mycelia of S. sclerotiorum during its in vitro germination. These metabolites were grouped based on their chemical composition and biological functions. Pathway analysis revealed all the enriched pathways which are discussed.
Identification and characterisation of secondary metabolites extracted from Sclerotinia sclerotiorum
Table 1 shows the chemical formula, chemical name, KEGG identification code, function, group, and subgroup of individual secondary metabolites detected in S. sclerotiorum in the current study.
| Table 1: Secondary metabolites produced by Sclerotinia sclerotiorum. | ||||||
| Formula | Metabolite name | Kegg Number | Function | Group | Subgroup | Chemical function |
| C4H9NO3 | L-homoserine (12647) | C00263 | Biological role | Peptide | Amino acids | Amino acid |
| C4H9NO3 | L-threonine (6288) | C00188 | Biological role | Peptide | Amino acids | Amino acid |
| C4H8N2O3 | Methylazoxymethanol acetate (5363199) | C19258 | Carcinogens | Group 2A carcinogenic compounds | Methyl ester, Azoxy compound | |
| C8H8O | Styrene oxide (7276) | C02083 | Carcinogens | Group 2A carcinogenic compounds | Epoxide | |
| C4H8N2O3 | N-nitroso-n-methylurethane (12001) | C19301 | Carcinogens | Group 2A carcinogenic compounds | Nitroso compound | |
| C21H40O3 | Glycidyl stearate (62642) | C19427 | Carcinogens | Group 3: Not carcinogenic to humans | Ester | |
| C3H6N2O | N-nitrosomethylvinylamine (20678) | C19282 | Carcinogens | Group 2A is probably carcinogenic to human compounds | Nitroso compound | |
| C29H48O2 | C11509 (443238) | C11509 | Lipids | Sterol lipids | Cholesterol and derivatives | Unknown |
| C29H50O2 | C04814 (440493) | C04814 | Lipids | Sterol lipids | Cholesterol and derivatives | Unknown |
| C31H48O3 | 3-hydroxy-vitamin k (5280540) | C02785 | Lipids | Quinones and hydroquinones | Vitamin K | Hydroxylated vitamin K |
| C5H11NO2 | 5-aminovaleric acid (138) | C00431 | Lipids | Fatty acyls | Amino fatty acids | Amino acid derivative |
| C18H28O3 | Alpha-licanic acid (5281118) | C08319 | Lipids | Fatty acyls | Oxo fatty acid | Carboxylic acid |
| C5H11NO2 | L-norvaline (65098) | C01826 | Lipids | Fatty acyls | Amino fatty acids | Amino acid |
| C15H11O7+ | Delphinidin (128853) | C05908 | Lipids | Flavonoids | Anthocyanidins | Flavonoid |
| C24H38O4 | C11637 (443323) | C11637 | Lipids | Sterol lipids | Bile acid, alcohols and derivatives | unknown compound |
| C40H54O | Echinenone (5281236) | C08592 | Lipids | Prenol lipids | Isoprenoids | Carotenoid |
| C13H18O2 | Plastoquinol-1 (24892729) | C02185 | Lipids | Quinones and hydroquinones | Ubiquinones | Quinone derivative |
| C18H28O3 | 12,13(s)-eotre (20843328) | C04672 | Lipids | Fatty acyls | Epoxy fatty acid | unknown |
| C40H56O2 | Deoxymyxol (16061292) | C15933 | Lipids | Prenol lipids | Isoprenoids | unknown |
| C40H54O | Hydroxychlorobactene (10099075) | C15911 | Lipids | Prenol lipids | Isoprenoids | unknown |
| C8H8O2 | 3-vinylcatechol (441226) | C07085 | Lipids | Octadecanoids | 12-oxophytodienoic acid metabolites | Vinyl-substituted catechol |
| C16H30O | Hexadecenal (5280541) | C06123 | Lipids | Fatty acyls | Fatty aldehydes | Aldehyde |
| C18H28O2 | Stearidonic acid (5312508) | C16300 | Lipids | Fatty acyls | Polyunsaturated fatty acids | Fatty acid |
| C18H28O3 | 9,10-eotre (23724711) | C16324 | Lipids | Fatty acyls | Other octadecanoids | Unknown |
| C31H48O3 | 2-hydroxy-vitamin k (11953813) | C02793 | Lipids | Quinones and hydroquinones | Vitamine K | Hydroxylated vitamin K |
| C13H2O3 | Methyl jasmonate (5281929) | C11512 | Lipids | Fatty acyls | Jasmonic acid | Methyl ester, Jasmonate |
| C18H28O3 | 10-opda (23724712) | C16325 | Lipids | Fatty acyls | Other octadecanoids | Unknown |
| C15H10O7 | 2'-hydroxypseudobaptigenin (5280616) | C03662 | Lipids | Flavonoids | Isoflavonoids | Hydroxylated flavonoid |
| C29H48O2 | (24r,28r)-fucosterol epoxide (440161) | C03910 | Lipids | sterol lipids | Stigmasterols | Epoxide |
| C17H14O6 | Aflatoxin b2 (2724360) | C16753 | Lipids, Mycotoxins | Polyketides, Aflatoxins | Aflatoxin and related substances | Mycotoxin |
| C29H50O2 | Alpha-tocopherol (14985) | C02477 | Lipids, Pharmaceutical additives in Japan, Japanese, OTC drugs risk category of Japanese OTC drugs | Quinones and hydroquinones, 3rd class OTC drugs, Nourishing tonics and health supplements | Vitamine E, Stabilizing agent | Vitamin E |
| C27H30O16 | Multinoside a (5319943) | C17563 | A major component of natural products | Crude drug | Glycoside | |
| C4H8N2O3 | Asparagine (236) | C16438 | A major component of natural products | Crude drug | Amino acid | |
| C5H11NO2 | Betaine (247) | C00719 | A major component of natural products | Crude drug | Quaternary ammonium compound | |
| C18H28O2 | Neoprene (6434236) | C19042 | Pesticides | Insect growth regulator | Juvenile hormone mimics | Insect growth regulator |
| C18H26O2 | Cinmethylin (91745) | C10903 | Pesticides | Herbicides | Herbicide | |
| C9H11NO2 | Metolcarb (14322) | C18747 | Pesticides, Target based compound | Insecticides, Enzyme | Inhibitor | Carbamate insecticide |
| C18H26O2 | Empenthrin (6434488) | C18524 | Pesticides, Target based compounds, Japanese Animal drugs | Insecticides, Ion channels, not therapeutic | Modulator | Insecticide |
| C5H14NO+ | Choline (305) | C00114 | Pesticides, Risk category of Japanese OTC drugs | Plant growth regulator, 3rd class OTC drugs | Inorganic and organic chemicals | Quaternary ammonium compound |
| C4H8N2O3 | Glycylglycine (11163) | C02037 | Pharmaceutical additives in Japan | Buffering agent | Dipeptide | |
| C15H10O4 | Chrysin (5281607) | C10028 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H12O5 | Naringenin (439246) | C00509 | Phytochemicals | Flavonoids | Flavanones | Flavonoid |
| C15H12O5 | Butein (5281222) | C08578 | Phytochemicals | Flavonoids | Chalcones | Flavonoid |
| C16H12O5 | Wogonin (5281703) | C10197 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C17H14O6 | Pinobanksin 3-o-acetate (148556) | C16418 | Phytochemicals | Flavonoids | Dihyroflavonols | Acetylated flavonoid |
| C21H2O12 | Bracteatin 6-o-glucoside (23724746) | C16410 | Phytochemicals | Flavonoids | Aurones | Glucosylated flavonoid |
| C16H12O5 | 3-methylgalangin (5281946) | C11577 | Phytochemicals | Flavonoids | Flavonols | Methylated flavonoid |
| C27H30O16 | Lucenin-2 (442615) | C10102 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H10O5 | 5-deoxykaempferol (5281611) | C10037 | Phytochemicals | Flavonoids | Flavonols | Flavonoid |
| C16H12O5 | 2'-hydroxyformononetin (5280551) | C02920 | Phytochemicals | Isoflavonoids | Isoflavones | Hydroxylated isoflavone |
| C15H10O5 | Morindone (442756) | C10376 | Phytochemicals | Polyketides | Anthraquinone | Unknown |
| C21H2O12 | Isoquercitrin (5280804) | C05623 | Phytochemicals | Flavonoids | Flavonols | Flavonoid, Glycoside |
| C15H10O4 | 4',6-dihydroxyflavone (182362) | C14344 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C16H12O5 | Acacetin (5280442) | C01470 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H12O5 | Pinobanksin (73202) | C09826 | Phytochemicals | Flavonoids | Dihyroflavonols | Flavonoid |
| C15H10O7 | Isoetin (5281649) | C10079 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H10O5 | Baicalein (5281605) | C10023 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C8H8O2 | 3,4-dihydroxystyrene (151398) | C06224 | Phytochemicals | Phenylpropanoids | Caffeate derivatives | Hydroxylated phenyl compound |
| C16H16O4 | Vestitol (177149) | C10540 | Phytochemicals | Isoflavonoids | Isoflavanes | Isoflavone |
| C15H12O4 | Aloe-emodin anthrone (122840) | C16760 | Phytochemicals | Polyketides | Anthrone | Anthraquinone derivative |
| C15H10O7 | Robinetin (5281692) | C10177 | Phytochemicals | Flavonoids | Flavonols | Flavonoid |
| C21H2O12 | Quercimeritrin (5282160) | C12639 | Phytochemicals | Flavonoids | Flavonols | Flavonoid, Glycoside |
| C15H10O4 | 7,4'-dihydroxyflavone (5282073) | C12123 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C16H16O4 | Perforatin a (441968) | C09014 | Phytochemicals | Pyrones | Chromoes | Unknown |
| C15H10O7 | 8-hydroxykaempferol (5280544) | C02806 | Phytochemicals | Flavonoids | Flavonols | Flavonoid |
| C29H48O3 | Messagenin (46173776) | C08631 | Phytochemicals | Triterpenoids | Dammarenes | Anthraquinone derivative |
| C15H11O5 | Luteolinidin (441701) | C08652 | Phytochemicals | Flavonoids | Anthocyanidins and anthocyanins | Flavonoid |
| C15H10O4 | 1,4-dihydroxy-2-methyl anthraquinone (99300) | C10329 | Phytochemicals | Polyketides | Anthraquinone | |
| C15H10O5 | 3',4',7-trihydroxy isoflavone (5284648) | C14313 | Phytochemicals | Isoflavonoids | Isoflavones | |
| C16H12O5 | Lucidin omega-methyl ether (149782) | C10370 | Phytochemicals | Polyketides | Anthraquinone | Methylated anthraquinone |
| C15H12O5 | Rubrofusarin (72537) | C09047 | Phytochemicals | Pyrones | Naphthopyrones | Unknown |
| C15H12O4 | Liquiritigenin (114829) | C09762 | Phytochemicals | Flavonoids | Flavanones | Flavonoid |
| C15H10O7 | Hypolaetin (5281648) | C10078 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H10O7 | Bracteatin (5281221) | C08577 | Phytochemicals | Flavonoids | Aurones | Flavonoid |
| C15H10O5 | Purpurin 1-methyl ether (442766) | C10397 | Phytochemicals | Polyketides | Anthraquinone | Methylated anthraquinone |
| C15H10O7 | Tricetin (5281701) | C10192 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H10O4 | Alizarin 2-methyl ether (80103) | C10291 | Phytochemicals | Polyketides | Anthraquinone | Flavonoid |
| C15H10O5 | Apigenin (5280443) | C01477 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C15H10O5 | Norwogonin (5281674) | C10113 | Phytochemicals | Flavonoids | Flavones | Flavonoid |
| C16H12O5 | Genkwanin (5281617) | C10046 | Phytochemicals | Flavonoids | Flavones | Unknown |
| C15H10O4 | Primetin (11055) | C10121 | Phytochemicals | Flavonoids | Flavones | Glycoside |
| C15H10O4 | Digiferrugineol (32209) | C10327 | Phytochemicals | Polyketides | Anthraquinone | Unknown |
| C27H30O16 | Rutin (5280805) | C05625 | Phytochemicals | Flavonoids | Flavonols | Anthraquinone derivative |
| C16H12O5 | Question (160717) | C01448 | Phytochemicals | Polyketides | Anthraquinone | Flavonoid |
| C15H10O5 | Emodin (3220) | C10343 | Phytochemicals | Polyketides | Anthraquinone | Flavonoid |
| C15H12O5 | 2,7,4'-trihydroxyisoflavanone (11954208) | C15567 | Phytochemicals | Isoflavonoids | Isoflavones | |
| C15H10O7 | Morin (5281670) | C10105 | Phytochemicals | Flavonoids | Flavonols | |
| C15H10O5 | Norobtusifolin (442759) | C10379 | Phytochemicals | Polyketides | Anthraquinone | |
| C15H10O4 | Rubiadin (124062) | C10402 | Phytochemicals | Polyketides | Anthraquinone | |
| C21H20O12 | Myricitrin (5281673) | C10108 | Phytochemicals | Flavonoids | Flavonols | |
| C15H12O5 | Chalconaringenin (5280960) | C06561 | Phytochemicals | Flavonoids | Chalcones | |
| C15H11O4 | Apigeninidin (441647) | C08574 | Phytochemicals | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H12O4 | Pinocembrin (68071) | C09827 | Phytochemicals | Flavonoids | Flavanones | |
| C27H30O16 | Sophoraflavonoloside (5282155) | C12634 | Phytochemicals | Flavonoids | Flavonols | |
| C15H12O4 | Pinocembrin chalcone (6474295) | C16404 | Phytochemicals | Flavonoids | Chalcones | |
| C16H12O5 | Physcion (10639) | C17045 | Phytochemicals | Polyketides | Anthraquinone | |
| C15H10O7 | 6-hydroxykaempferol (5281638) | C10068 | Phytochemicals | Flavonoids | Flavonols | |
| C16H16O4 | 2',6'-dihydroxy-4'-methoxydihydrochalcone (169676) | C09644 | Phytochemicals | Flavonoids | Dihydrochalcones | |
| C15H12O4 | 3,9-dihydroxypterocarpan (162933) | C04271 | Phytochemicals | Isoflavonoids | Pterocarpans | |
| C17H14O6 | Ventinone a (442767) | C10407 | Phytochemicals | Polyketides | Anthraquinone | |
| C16H12O5 | Melanin (442808) | C10504 | Phytochemicals | others | Neoflavonoids | |
| C15H12O5 | Garbanzol (442410) | C09751 | Phytochemicals | Flavonoids | Dihyroflavonols | |
| C15H12O4 | Hydrangenol (119199) | C10262 | Phytochemicals | Skimate / acetate-malonate pathway-derived compounds | Miscellaneous stilbenoids | |
| C27H30O17 | Baimaside (5282166) | C12667 | Phytochemicals | Flavonoids | Flavonols | |
| C15H10O7 | 6-hydroxyluteolin (5281642) | C10072 | Phytochemicals | Flavonoids | Flavones | |
| C21H20O12 | Gossypetin 8-rhamnoside (5281620) | C10050 | Phytochemicals | Flavonoids | Flavonols | |
| C15H10O7 | Quercetin (5280343) | C00389 | Phytochemicals, Carcinogens, transporter | Flavonoids, Group 3-not carcinogenic to humans, Solute carrier | Flavonols, Inhibitor | |
| C24H38O4 | Dioctyl phthalate (8343) | C03690 | Phytochemicals, Carcinogens, Endocrine disrupting compound | Polyketides, Group 2B, possibly carcinogenic to humans compounds, Plasticizers, and plastics | Anthraquinone, Phthalates | |
| C17H15O6 | Rosinidin (441777) | C08729 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H11O5 | Pelargonidin (440832) | C05904 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H10O4 | Daidzein (5281708) | C10208 | Phytochemicals, lipids | Flavonoids | Isoflavones | |
| C40H56O2 | Zeaxanthin (5280899) | C06098 | Phytochemicals, lipids | Carotenoids, apocarotenoids, and prenol lipids | Carotenoids, isoprenoids | |
| C15H12O5 | 2'-hydroxydihydrodaidzein (440047) | C03567 | Phytochemicals, lipids | Flavonoids | Flavones | |
| C17H14O6 | Pisatin (101689) | C10516 | Phytochemicals, lipids | Isoflavonoids | Pterocarpans | |
| C16H12O5 | Inermin (91510) | C10502 | Phytochemicals, lipids | Isoflavonoids | Pterocarpans | |
| C27H31O16 | Cyanidin 3-o-sophoroside (11169452) | C16306 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H12O4 | Isoliquiritigenin (638278) | C08650 | Phytochemicals, lipids | Flavonoids | Chalcones | |
| C16H12O5 | Prunetin (5281804) | C10521 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C15H10O4 | His idol (5281254) | C08644 | Phytochemicals, lipids | Flavonoids | Aurones | |
| C21H21O12 | Mirtillin (443650) | C12138 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C40H56O2 | Lactucaxanthin (5281242) | C08599 | Phytochemicals, lipids | Carotenoids, apocarotenoids, and prenol lipids | Carotenoids, isoprenoids | |
| C15H10O4 | Anhydroglycinol (442667) | C10200 | Phytochemicals, lipids | Isoflavonoids | Pterocarpans | |
| C26H28O16 | C12637 (5487635) | C12637 | Phytochemicals, lipids | Isoflavonoids | Pterocarpans | |
| C16H12O5 | Biochanin a (5280373) | C00814 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C16H12O5 | Calycosin (5280448) | C01562 | Phytochemicals, lipids | Isoflavonoids, a crude drug | Isoflavones | |
| C16H12O5 | Texasin (5281812) | C10536 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C17H14O6 | Irisolidone (5281781) | C10471 | Phytochemicals, lipids | Flavonoids | Isoflavones | |
| C27H31O17 | Delphin (10100906) | C16312 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H10O5 | 2'-hydroxydaidzein (5280520) | C02495 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C15H10O5 | Genistein (5280961) | C06563 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C16H12O5 | Glycitein (5317750) | C14536 | Phytochemicals, lipids | Isoflavonoids | Isoflavones | |
| C15H10O5 | Aloe-emodin (10207) | C10294 | Phytochemicals, lipids | Polyketides | Anthraquinone | |
| C27H31O17+ | Delphinidin 3-o-sophoroside (23724705) | C16307 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C27H31O16+ | Tulipanin (5492231) | C16315 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C15H10O4 | Chrysophanol (10208) | C10315 | Phytochemicals, a major component of natural products | Polyketides, crude drug | Anthraquinone | |
| C16H12O5 | Obtusifolin (3083575) | C17039 | Phytochemicals, a major component of natural products | Polyketides | Anthraquinone | |
| C37H40O9 | Resiniferatoxin (442082) | C09179 | Phytochemicals, target-based compounds | Terpenoids, voltage-dated cations channels | Daphnanes, Agonist | |
| C40H56O2 | Lutein (5281243) | C08601 | Phytochemicals, lipids | Carotenoids, apocarotenoids, and prenol lipids | Carotenoids, isoprenoids | |
| C27H31O16+ | Cyanin (441688) | C08639 | Phytochemicals, lipids | Flavonoids | Anthocyanidins and anthocyanins | |
| C20H23N5O6S | Azlocillin (6479523) | C06839 | Unclassified | |||
| C9H11NO2 | 4-hydroxy-1-(3-pyridinyl)-1-butanone (107819) | C19565 | Unclassified | |||
| C8H8O2 | Benzylformate (7708) | C05613 | Unclassified | |||
| C24H38O4 | Diisooctyl phthalate (33934) | C14577 | Unclassified | |||
| C8H14O5S | 2-(3'-methylthio)propyl malate (24883455) | C17214 | Unclassified | |||
| C8H8O2 | 2-methyl benzoic acid (8373) | C07215 | Unclassified | |||
| C9H11NO2 | Tricaine (11400) | C18090 | Unclassified | |||
| C24H38O4 | Di-n-octyl phthalate (8346) | C14227 | Unclassified | |||
| C9H11NO2 | Benzocaine (2337) | C07527 | Unclassified | |||
| C17H14O6 | 4',6-dihydroxy-5,7-dimethoxyflavone (244386) | C15100 | Unclassified | |||
| C4H9NO3 | 2-methylserine (439656) | C02115 | Unclassified | |||
| C15H12O5 | P-coumaroyltriacetic acid lactone (54704424) | C12087 | Unclassified | |||
| C15H10O5 | Sulfuretin (5281295) | C08730 | Unclassified | |||
| C18H28O3 | Etherolenic acid (23724709) | C16319 | Unclassified | |||
| C35H36N4O5 | Pheophorbide a (5323510) | C18021 | Unclassified | |||
| C8H8O2 | Phenylacetate (31229) | C00548 | Unclassified | |||
| C16H16O4 | 1,2-bis(4-hydroxy-3-methoxyphenyl)ethylene (5280698) | C04547 | Unclassified | |||
| C16H16O4 | 9-methoxy-alpha-lapachone (442754) | C10372 | Unclassified | |||
| C15H12O5 | Licodione (439528) | C01592 | Unclassified | |||
| C15H10O5 | 6-hydroxydaidzein (5284649) | C14314 | Unclassified | |||
| C18H28O3 | 17beta-hydroxy-2-oxa-5alpha-androstane-3-one (252289) | C14911 | Unclassified | |||
| C18H26O2 | Prenortestosterone (235672) | C15257 | Unclassified | |||
| C8H8O | 3-methylbenzaldehyde (12105) | C07209 | Unclassified | |||
| C21H20O12 | 6-hydroxy luteolin 7-glucoside (185766) | C17763 | Unclassified | |||
| C8H8O2 | Methyl benzoate (7150) | C20645 | Unclassified | |||
| C16H16O4 | Eleutherin (10166) | C10340 | Unclassified | |||
| C15H12O5 | Toralactone (5321980) | C17673 | Unclassified | |||
| C24H38O4 | Apocholic acid (101818) | C15375 | Unclassified | |||
| C27H31O16+ | Cyanidin 3,7-di-o-beta-d-glucoside (5491675) | C20469 | Unclassified | |||
| C47H70O3 | 2-octaprenyl-6-methoxy-1,4-benzoquinone (5280835) | C05813 | Unclassified | |||
| C8H8O2 | M-toluic acid (7418) | C07211 | Unclassified | |||
| C7H14N2O6S | Glutaurine (68759) | C05844 | Unclassified | |||
| C9H11NO2 | 5-(3-pyridyl)-2-hydroxytetrahydrofuran (179630) | C19578 | Unclassified | |||
| C8H21NO6P+ | Glycerophosphocholine (439285) | C00670 | Unclassified | |||
| C47H70O3 | 3-octa prenyl-4-hydroxybenzoate (5280831) | C05809 | Unclassified | |||
| C8H8O2 | 4-hydroxyphenyl acetaldehyde (440113) | C03765 | Unclassified | |||
| C15H10O5 | Lucidin (10163) | C10369 | Unclassified | |||
| C5H11NO2 | Valine (1182) | C16436 | Unclassified | |||
| C18H26O2 | Nandrolone (9904) | C07254 | Unclassified | |||
| C8H8O2 | Phenylacetic acid (999) | C07086 | Unclassified | |||
| C40H56O2 | Rhodopinal (20055178) | C16270 | Unclassified | |||
| C27H31O17+ | Delphinidin 3,7-di-o-beta-d-glucoside (72734296) | C20496 | Unclassified | |||
| C15H10O5 | Islandicin (10151) | C16796 | Unclassified | |||
| C8H8O | P-tolu aldehyde (7725) | C06758 | Unclassified | |||
| C13H20O3 | (6s,9r)-vomifoliol (5280462) | C01760 | Unclassified | |||
| C29H48O3 | C04840 (440507) | C04840 | Unclassified | |||
| C13H18O2 | Ibuprofen (3672) | C01588 | Unclassified | |||
| C8H14O5S | 3-(3'-methylthio)propyl malate (44237293) | C17215 | Unclassified | |||
| C8H8O | Phenylacetaldehyde (998) | C00601 | Unclassified | |||
| C5H9NO2 | Proline (614) | C16435 | Unclassified | |||
| C17H14O6 | Cirsimaritin (188323) | C17785 | Unclassified | |||
| C8H8O2 | 4'-hydroxyacetophenone (7469) | C10700 | Unclassified | |||
| C8H8O2 | P-anisaldehyde (31244) | C10761 | Unclassified | |||
| C8H8O | 2-methylbenzaldehyde (10722) | C07214 | Unclassified | |||
| C15H11O7+ | 6-hydroxycyanidin (441697) | C08646 | Unclassified | |||
| C16H12O5 | Geraldine (5281618) | C10047 | Unclassified | |||
| C4H8N2O3 | N-carbamoylsarcosine (439375) | C01043 | Unclassified | |||
| C8H8O2 | 3-methylsalicylaldehyde (522777) | C14087 | Unclassified | |||
| C8H8O | Acetophenone (7410) | C07113 | Unclassified | |||
| C9H11NO2 | Phenylalanine (994) | C02057 | Unclassified | |||
| C15H10O5 | Galangin (5281616) | C10044 | Unclassified | |||
| C5H11NO2 | 4-methylaminobutyrate (70703) | C15987 | Unclassified | |||
| C15H12O5 | Dihydrogenistein (9838356) | C14458 | Unclassified | |||
| C4H8N2O3 | 3-ureidopropionate (111) | C02642 | Unclassified | |||
| C15H12O5 | Butin (92775) | C09614 | Unclassified | |||
| C16H12O5 | Cypripedium (174864) | C10323 | Unclassified | |||
| C35H52O4 | Hyperforin (441298) | C07608 | Unclassified | |||
| C5H11NO2 | 2-amino-2-methyl butanoate (94744) | C03571 | Unclassified | |||
| C15H10O5 | 3,6,4'-trihydroxy flavone (676308) | C15222 | Unclassified | |||
| C15H12O4 | 3',5'-dihydroxy flavanone (11954216) | C15609 | Unclassified | |||
| C17H14O6 | 4'-methylcapillarisin (5320438) | C17784 | Unclassified | |||
| C16H30O | Bombykol (445128) | C16873 | Unclassified | |||
| C31H48O3 | Dehydroeburicoic acid (15250826) | C16950 | Unclassified | |||
| C17H14O6 | Aflatoxicol (53297443) | C19584 | Unclassified | |||
| C8H8O2 | 4-methyl benzoic acid (7470) | C01454 | Unclassified | |||
| C15H10O7 | Nortangeretin (96506) | C15031 | Unclassified | |||
| C8H8O | 4-vinylphenol (62453) | C05627 | Unclassified | |||
| C15H12O5 | (-)-Glycinol (129648) | C01263 | Unclassified | |||
| C4H9NO3 | (-)-Erythro-(2r,3r)-dihydroxybutylamide (443073) | C11108 | Unclassified | |||
| C18H28O3 | Colnelenic acid (6441679) | C16320 | Unclassified | |||
| C5H9NO2 | 3-acetamidopropanal (5460495) | C18170 | Unclassified | |||
| C27H30O16 | Quercetin 3-o-rhamnoside 7-o-glucoside (6325870) | C19796 | Unclassified | |||
| C26H29O16+ | Delphinidin 3-o-beta-d-sambubioside (10196837) | C20491 | Unclassified | |||
| C4H9NO3 | Gabob (2149) | C03678 | Unclassified | |||
| C5H11NO2 | Isoamyl nitrite (8053) | C07457 | Unclassified | |||
| C8H8O2 | 2-hydroxyacetophenone (68490) | C07189 | Unclassified | |||
| C16H16O4 | Deoxyshikonin (98914) | C18133 | Unclassified | |||
| C15H12O5 | 6,7,4'-trihydroxyflavanone (23724670) | C16232 | Unclassified | |||
| C18H28O3 | 12-opda (5280411) | C01226 | Unclassified | |||
| C15H12O4 | Cis-3,4-phenanthrenedihydrodiol-4-carboxylate (49787035) | C18256 | Unclassified | |||
| C9H11NO2 | L-beta-phenylalanine (686703) | C20487 | Unclassified | |||
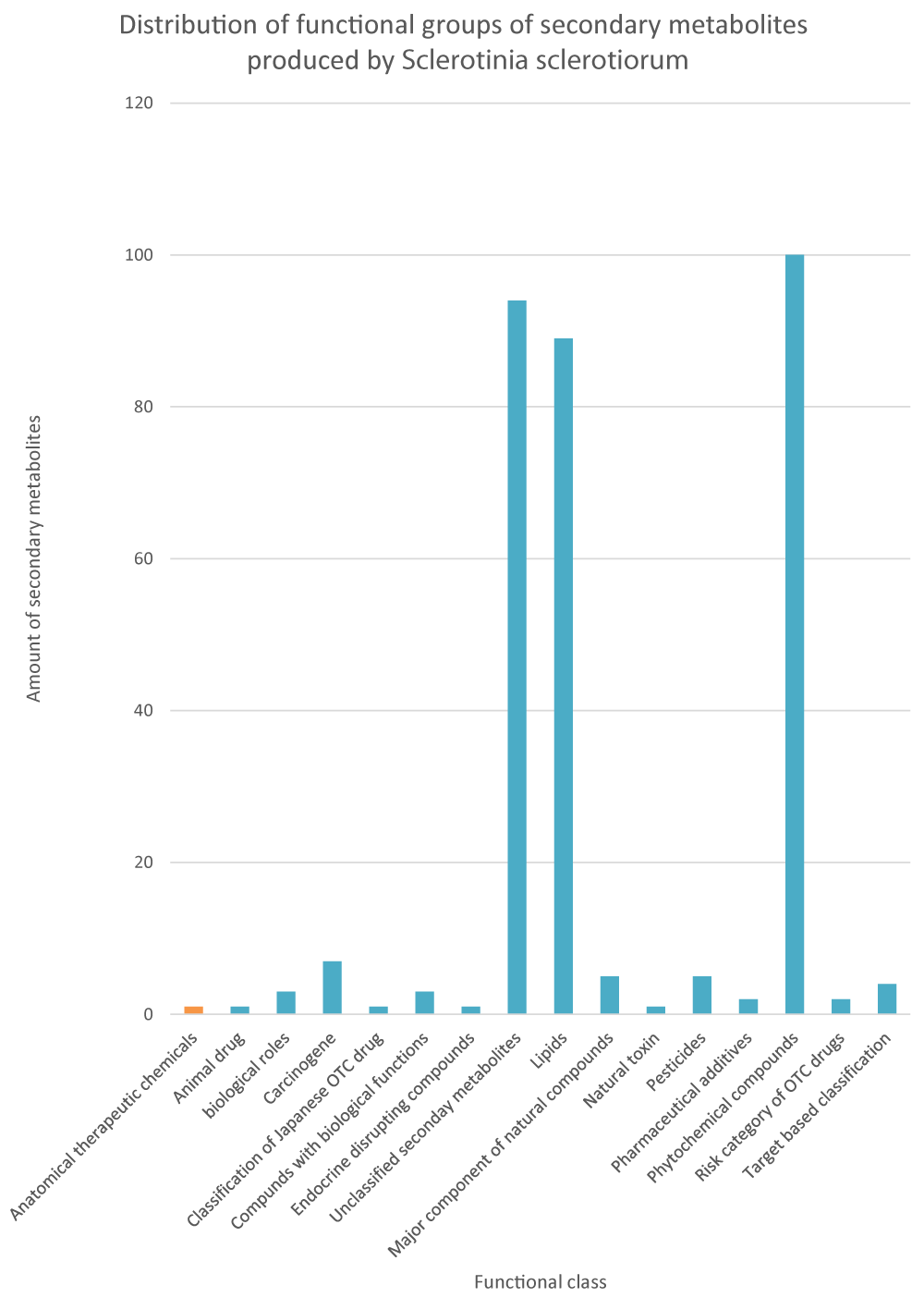
The distribution of functional classification of secondary metabolites produced by S. sclerotiorum is represented in Figure 2. The secondary metabolites profile shows that S. sclerotiorum has; phytochemical compounds (100), lipids (89), carcinogens (7), pesticides (5), major components of natural products (5), target-based classification of chemical compounds (4), chemical compounds with biological roles (3), pharmaceutical additives in Japan (2), risk category of Japanese otc drugs (2), natural toxins (1), animal drugs in Japan (1), classification of Japanese otc drugs (1), endocrine disrupting compounds (1) and 94 unclassified secondary metabolites.
Figure 2: Functional classification of secondary metabolites produced by Sclerotinia sclerotiorum, based on the annotation from the KEGG database.
Pathway analysis of secondary metabolites produced by Sclerotinia sclerotiorum
The pathway analysis results of all the secondary metabolites detected in this investigation were represented graphically in Figure 3 and Table 2 to simplify the biological implication of the enriched pathways connected with the identified secondary metabolites.
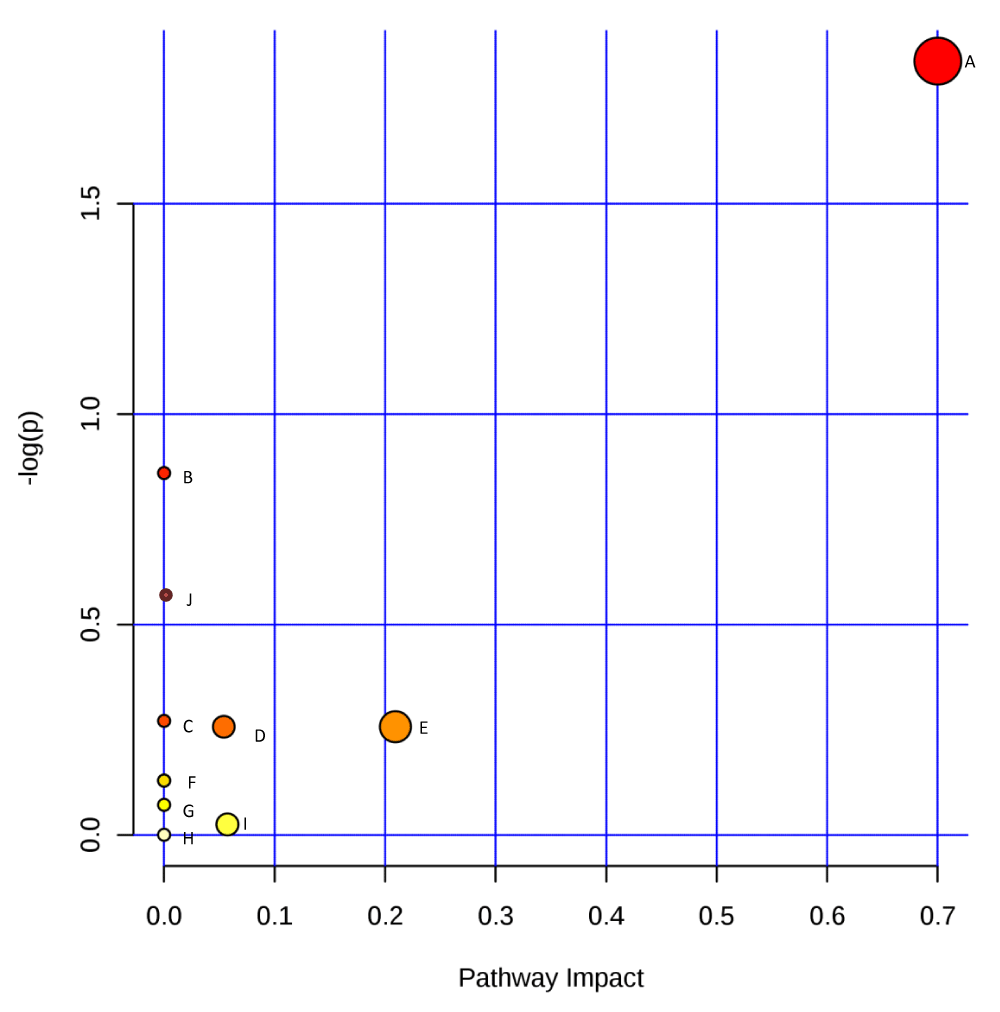
Figure 3: Graphical representation of impact value of enriched pathways associated with secondary metabolites produced by Sclerotinia sclerotiorum. –log p is the log value of the original p value calculated from the enrichment analysis; pathway impact is the impact value calculated from pathway topology analysis. A (Phenylalanine metabolism); B (Taurine and hypotaurine metabolism); C (Sulfur metabolism); D (Glycerophospholipid metabolism); E (Glycine, serine, and threonine metabolism); F (Lysine biosynthesis); G (Tyrosine metabolism); H (Valine, leucine and isoleucine biosynthesis); I (Cysteine and methionine metabolism); J (Aminoacyl-tRNA biosynthesis).
| Table 2: Pathway Analysis of secondary metabolites produced by Sclerotinia sclerotiorum. | ||||||||
| Pathway | Total | Expected | Hits | Raw p | -log(p) | Holm adjust P | FDR | Impact |
| Phenylalanine metabolism | 7 | 0.73 | 2 | 1.59E-01 | 1.84E+00 | 1.00E+00 | 1.00E+00 | 0.70 |
| Taurine and hypotaurine metabolism | 5 | 0.52 | 1 | 4.23E-01 | 8.60E-01 | 1.00E+00 | 1.00E+00 | 0.00 |
| Sulfur metabolism | 13 | 1.35 | 1 | 7.63E-01 | 2.71E-01 | 1.00E+00 | 1.00E+00 | 0.00 |
| Glycerophospholipid metabolism | 26 | 2.70 | 2 | 7.73E-01 | 2.57E-01 | 1.00E+00 | 1.00E+00 | 0.05 |
| Glycine, serine, and threonine metabolism | 26 | 2.70 | 2 | 7.73E-01 | 2.57E-01 | 1.00E+00 | 1.00E+00 | 0.21 |
| Lysine biosynthesis | 19 | 1.98 | 1 | 8.79E-01 | 1.29E-01 | 1.00E+00 | 1.00E+00 | 0.00 |
| Tyrosine metabolism | 19 | 1.98 | 1 | 8.79E-01 | 1.29E-01 | 1.00E+00 | 1.00E+00 | 0.00 |
| Valine, leucine, and isoleucine biosynthesis | 24 | 2.50 | 1 | 9.31E-01 | 7.15E-02 | 1.00E+00 | 1.00E+00 | 0.00 |
| Cysteine and methionine metabolism | 33 | 3.43 | 1 | 9.75E-01 | 2.51E-02 | 1.00E+00 | 1.00E+00 | 0.06 |
| Aminoacyl-tRNA biosynthesis | 67 | 6.97 | 1 | 1.00E+00 | 4.65E-04 | 1.00E+00 | 1.00E+00 | 0.00 |
| In particular, the Total is the total number of metabolites in the pathway; the Hits is the matched number of metabolites from the current study; Raw p is the p - value calculated from the enrichment analysis; the Holm p is the p - value adjusted by the Holm-Bonferroni method; the FDR p is the p - value adjusted using False Discovery Rate; Impact is the pathway impact value calculated from pathway topology analysis. | ||||||||
Figure 3 distinctively identified the phenylalanine metabolism pathway as the most significant pathway with an impact value of 0.7, followed by glycine, serine, and threonine metabolism pathways having 0.2 as an impact value; however, other identified pathways have negligible impact values.
Although significant advancement has been made in understanding the molecular characteristics of the necrotrophic plant pathogen- S. sclerotiorum, several aspects of its lifecycle and infection processes remain vague [22].
This research is a progression from these fungi's genomic, transcriptomic, and proteomic analysis. While transcriptomics studies generated essential data relating to S. sclerotiorum gene expression during in vitro growth stage [23], proteomic results justified the transcriptomic results with the list of the corresponding protein [7] and the secondary metabolites profile gave a clear picture of the outcome of the cellular processes that occur within S. sclerotiorum. This research profiled the secondary metabolites produced by S. sclerotiorum during in vitro germination, revealing this fungi's richness.
S. sclerotiorum, like many other phytopathogenic fungi, is a biosynthetically endowed organism that produces a massive range of chemically diverse and biologically significant molecules known as metabolites. Nonetheless, the S. sclerotiorum metabolome profiling conducted in the current study revealed a catalog of secondary metabolites produced by S. sclerotiorum during in vitro germination is discussed below.
Sclerotinia sclerotiorum produces a plethora of diverse and bioactive secondary metabolites
Secondary metabolites produced by S. sclerotiorum, identified in this study, were classified as phytochemicals, lipids, and natural toxins based on their chemical constituents and known functions (Table 1).
Phytochemical compounds: In the current study, 100 phytochemical compounds were identified as part of S. sclerotiorum secondary metabolites. These phytochemicals were classified into sub-groups: alkaloids, amino acid-associated compounds, flavonoids, fatty acids-related compounds, phenylpropanoids, polyketides, skate/acetate-malonate pathway derivative compounds, and terpenoids.
Moreover, several identified secondary metabolites belong to the flavonoid group, which could indicate the antimicrobial properties inert within S. sclerotiorum [24]. As shown in Table 1, the results show that S. sclerotiorum produces more flavonoids as secondary metabolites, although the function of these flavonoids in S. sclerotiorum is still largely unknown. However, according to [25], a similar ALT1 ligand was identified as a methylated flavonoid produced by Alternaria spp associated with Asthma in humans.
Lipids: In the metabolome of S. sclerotiorum, 89 lipid compounds were identified as part of its secondary metabolites. These lipids belong to eight classes, including fatty acyls, glycerolipids, glycerophospholipid, sphingolipids, sterol lipids, prenol lipids, saccharolipids, polyketides, exhibiting varying functions, including energy storing and acting as structural components of cell membranes [26]. For instance, 18 polyketides (molindone, aloe emodin anthrone, and 1,4-Dihydroxy-2-methylanthraquinone) were secreted by S. sclerotiorum, yet their mechanism of action is still elusive. However, studies have demonstrated that many polyketides, whose backbones are often frequently changed by glycosylation or oxidation, e.g., erythromycins, tetracyclines, and avermectins, are commonly utilized antimicrobial, anti-parasitic, and anti-cancer and antitumor compounds [27,28]. Five of the lipids identified in S. sclerotiorum were reported to belong to the alpha-linolenic acid metabolism pathway, and eight others were implicated in the biosynthesis of the secondary metabolites pathway.
Natural toxins: Natural toxins include fungal toxins (mycotoxin), phytotoxins, cyanotoxins, marine biotoxins, and venoms. Aflatoxin B2 and resiniferatoxin (identified in the current study) are toxins produced by S. sclerotiorum as this collaborated with the report of [29], wherein they identified S. sclerotiorum P450 enzymes that are associated with aflatoxin biosynthetic pathway.
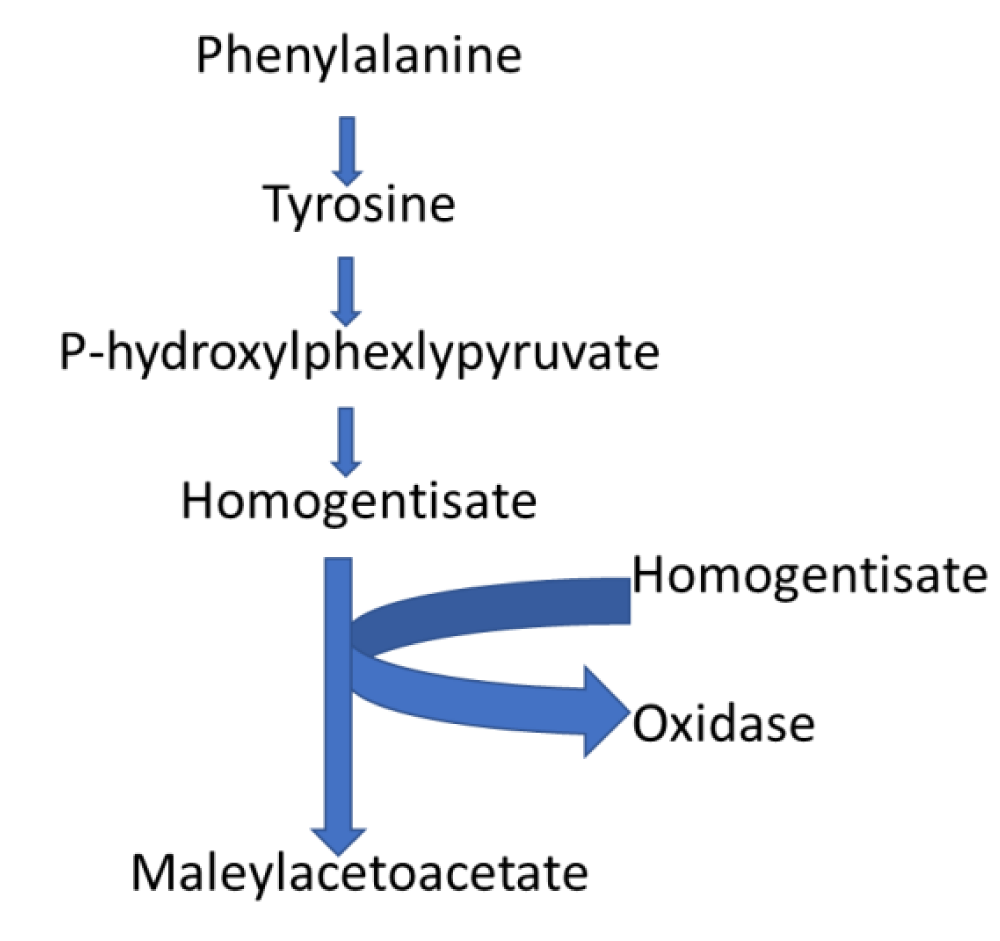
Impact of phenylalanine metabolism in the biochemical pathways associated with Sclerotinia sclerotiorum: Pathway analysis results identified 2-phenylacetamide, phenylacetic acid, phenylacetaldehyde, phenyl pyruvic acid, and L-phenylalanine secondary metabolites involved in the phenylalanine metabolism pathway that was enriched compared to other detected pathways (Figure 4). Although the significance of this pathway in S. sclerotiorum is still vague, it was reported that reprogramming of the phenylalanine cycle is responsible for soybean resistance against S. sclerotiorum attack [30]. Phenylpyruvic acid is a pyruvate dehydrogenase inhibitor essential for the metabolism of glucose, fatty acids, and cholesterol [31]. Likewise, phenylacetaldehyde is responsible for polyesters synthesis, managing additive activities during the polymerization process [32], while phenylacetic acid is a nitrogen and ammonium binding agent [33]. Based on the known functions of these individual secondary metabolites implicated in the phenylalanine pathway, it could be proposed that; Phenylalanine metabolism pathway is responsible for inhibiting the host plant phenylalanine defense mechanism [34,35].
Figure 4: Phenylalanine metabolism pathway, the most significant pathway implicated with the secondary metabolism of Sclerotinia sclerotiorum.
Limitations of the study: The study on the secondary metabolites profiling of the phytopathogenic fungus Sclerotinia sclerotiorum has several limitations. Firstly, the research may have only focused on a specific strain or isolates of S. sclerotiorum, which may limit the generalizability of the findings to other strains or species [36]. The study might have included a partial analysis of all possible secondary metabolites produced by the fungus, as detecting and identifying secondary metabolites can be challenging and dependent on the analytical techniques employed [37]. Moreover, the study may have been conducted under specific laboratory conditions, which may not fully represent the natural environment in which the fungus interacts with plants [38]. Additionally, the functional characterization of the identified secondary metabolites and their role in pathogenicity may require further investigation [39]. These limitations should be taken into consideration when interpreting and extrapolating the results of the study.
In conclusion, the study on the secondary metabolites profiling of the phytopathogenic fungus Sclerotinia sclerotiorum revealed significant insights into its chemical composition. The research successfully identified and characterized several secondary metabolites produced by S. sclerotiorum, providing valuable information about its bioactive compounds and their potential role in pathogenicity. Two hundred and forty metabolites were found to vary in abundance between biological replicates. The metabolites included essential groups of compounds such as phytochemicals, lipids, and toxins, amongst others. Many of these metabolites were involved in critical pathways associated with resistance, nitrogen remobilization, cell signaling, and secondary metabolic defenses [40]. Metabolites discovered in this research are potentially primarily related to the production of secondary metabolites, indicating the level of all housekeeping metabolites since the pathogen was grown in vitro, excluding the metabolites expressed during the pathogenicity of host plants.
In summary, these data support that both secondary metabolites are involved in multiple interconnecting pathways that contribute immensely to the pathogenicity of S. sclerotiorum.
These findings contribute to our understanding of S. sclerotiorum's chemical arsenal and offer potential targets for disease management strategies. However, further investigations are needed to fully comprehend the functional significance of these secondary metabolites and their interactions with host plants. Future studies could focus on elucidating the mechanisms underlying the fungus-host interactions and exploring the potential application of these metabolites in agricultural practices. Such research holds promise for developing innovative approaches to combating plant diseases caused by S. sclerotiorum [41].
This work was supported by the South African National Research Foundation (NRF; No. TTK170413227119) and the SARChI program of the Department of Science and Technology and the Wits Research Office for post-doctoral fellowship funding.
- Purdy LH. Symposium on Sclerotinia sclerotiorum: History, Diseases and Symptomatology, Host Range, Geographic Distribution, and Impact. Phytopathology. 1979; 69(8):875–80.
- Rahman MME, Suzuki K, Islam MM, Dey TK, Harada N, Hossain DM. Molecular characterisation, mycelial compatibility grouping, and aggressiveness of a newly emerging phytopathogen, Sclerotinia sclerotiorum, causing white mould disease in new host crops in Bangladesh. Journal of Plant Pathology. 2020; 102(3):775–85.
- Gupta T, Kumari C, Vanshika, Kulshrestha S. Biology and mycovirus-assisted biological control of Sclerotinia sclerotiorum infecting vegetable and oilseed crops. Archives of Phytopathology and Plant Protection. 2019; 52(13-14):1049–67.
- Hossain MM, Sultana F, Li W, Tran LP, Mostofa MG. Sclerotinia sclerotiorum (Lib.) de Bary: Insights into the Pathogenomic Features of a Global Pathogen. Cells. 2023 Mar 31;12(7):1063. doi: 10.3390/cells12071063. PMID: 37048136; PMCID: PMC10093061.
- Tian J, Chen C, Sun H, Wang Z, Steinkellner S, Feng J, Liang Y. Proteomic Analysis Reveals the Importance of Exudates on Sclerotial Development in Sclerotinia sclerotiorum. J Agric Food Chem. 2021 Feb 3;69(4):1430-1440. doi: 10.1021/acs.jafc.0c06685. Epub 2021 Jan 22. PMID: 33481591.
- Otun S, Ntushelo K. How to Knock down a Plant; the Three Weapons of Sclerotinia sclerotiorum. Journal of Biological Sciences. 2019; 19(4):300–13.
- Otun S, Ntushelo K. Proteomic analysis of the phytogenic fungus Sclerotinia sclerotiorum. J Chromatogr B Analyt Technol Biomed Life Sci. 2020 May 1;1144:122053. doi: 10.1016/j.jchromb.2020.122053. Epub 2020 Mar 14. PMID: 32229427.
- Zamani-Noor N. Baseline Sensitivity and Control Efficacy of Various Groups of Fungicides against Sclerotinia sclerotiorum in Oilseed Rape Cultivation. Agronomy. 2021; 11(9):1758.
- Albert D, Dumonceaux T, Carisse O, Beaulieu C, Filion M. Combining Desirable Traits for a Good Biocontrol Strategy against Sclerotinia sclerotiorum. Microorganisms. 2022 Jun 9;10(6):1189. doi: 10.3390/microorganisms10061189. PMID: 35744707; PMCID: PMC9228387.
- Verma PP, Shelake RM, Das S, Sharma P, Kim JY. Plant Growth-Promoting Rhizobacteria (PGPR) and Fungi (PGPF): Potential Biological Control Agents of Diseases and Pests. In: Microbial Interventions in Agriculture and Environment. Singapore: Springer Singapore. 2019; 281–311.
- Singh Saharan G, Mehta NK, Meena PD. Future Research Priorities of Crucifers’ Host-Pathosystem. In: Genomics of Crucifer’s Host- Pathosystem. Singapore: Springer Nature Singapore. 2023; 915–21.
- Tian J, Chen C, Sun H, Wang Z, Steinkellner S, Feng J. Proteomic Analysis Reveals the Importance of Exudates on Sclerotial Development in Sclerotinia sclerotiorum. J Agric Food Chem [Internet]. 2021 Feb 3 [cited 2023 Jan 12];69(4):1430–40. https://pubs.acs.org/doi/full/10.1021/acs.jafc.0c06685
- Zuo R, Xie M, Gao F, Sumbal W, Cheng X, Liu Y, Bai Z, Liu S. The Characterization of the Phloem Protein 2 Gene Family Associated with Resistance to Sclerotinia sclerotiorum in Brassica napus. Int J Mol Sci. 2022 Apr 1;23(7):3934. doi: 10.3390/ijms23073934. PMID: 35409295; PMCID: PMC8999561.
- Li M, Tang Y, Yu M, Fan Y, Khan SU, Chang W. Systematic Characterisation of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance. Horticulturae [Internet]. 2022 Oct 1 [cited 2023 Jan 12];8(10):874. https://www.mdpi.com/2311-7524/8/10/874/htm
- Thaning C, Welch CJ, Borowicz JJ, Hedman R, Gerhardson B. Suppression of Sclerotinia sclerotiorum apothecial formation by the soil bacterium Serratia plymuthica: identification of a chlorinated macrolide as one of the causal agents. Soil Biol Biochem. 2001; 33(12–13):1817–26.
- Li H, Cai Y, Guo Y, Chen F, Zhu ZJ. MetDIA: Targeted Metabolite Extraction of Multiplexed MS/MS Spectra Generated by Data-Independent Acquisition. Anal Chem. 2016 Sep 6;88(17):8757-64. doi: 10.1021/acs.analchem.6b02122. Epub 2016 Aug 8. PMID: 27462997.
- Bang KW. Investigation of plant and fungi-derived natural products for application as antimicrobial aerosols. 2020 [cited 2023 Jan 12]; https://researchspace.auckland.ac.nz/handle/2292/54253
- Zhang J, Lu Q, Xin L, Lou Y, Xiao W, Wang Z, Zhao L, Xiong Z. A liquid chromatography-mass spectrometry untargeted urinary metabonomics combined with quantitative analysis of seven amino acids biomarkers on yaobitong capsule in the intervention of rheumatoid arthritis rats. J Sep Sci. 2022 Dec;45(23):4209-4223. doi: 10.1002/jssc.202200654. Epub 2022 Oct 17. PMID: 36200630.
- Ridder L, van der Hooft JJ, Verhoeven S. Automatic Compound Annotation from Mass Spectrometry Data Using MAGMa. Mass Spectrom (Tokyo). 2014;3(Spec Iss 2):S0033. doi: 10.5702/massspectrometry.S0033. Epub 2014 Jul 2. PMID: 26819876; PMCID: PMC4321337.
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008 Jan;36(Database issue):D480-4. doi: 10.1093/nar/gkm882. Epub 2007 Dec 12. PMID: 18077471; PMCID: PMC2238879.
- Chong J, Wishart DS, Xia J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr Protoc Bioinformatics. 2019 Dec;68(1):e86. doi: 10.1002/cpbi.86. PMID: 31756036.
- Xia S, Xu Y, Hoy R, Zhang J, Qin L, Li X. The Notorious Soilborne Pathogenic Fungus Sclerotinia sclerotiorum: An Update on Genes Studied with Mutant Analysis. Pathogens. 2019 Dec 27;9(1):27. doi: 10.3390/pathogens9010027. PMID: 31892134; PMCID: PMC7168625.
- Maximiano MR, Miranda VJ, de Barros EG, Dias SC. Validation of an in vitro system to trigger changes in the gene expression of effectors of Sclerotinia sclerotiorum. J Appl Microbiol. 2021 Aug;131(2):885-897. doi: 10.1111/jam.14973. Epub 2021 Jan 4. PMID: 33331046.
- Can H, Kal U, Kayak N, Dal Y, Turkmen O. Use of microbial inoculants against biotic stress in vegetable crops: physiological and molecular aspect. In: Sustainable Horticulture. Elsevier; 2022; 263–332.
- Garrido-Arandia M, Silva-Navas J, Ramírez-Castillejo C, Cubells-Baeza N, Gómez-Casado C, Barber D, Pozo JC, Melendi PG, Pacios LF, Díaz-Perales A. Characterisation of a flavonoid ligand of the fungal protein Alt a 1. Sci Rep. 2016 Sep 16;6:33468. doi: 10.1038/srep33468. PMID: 27633190; PMCID: PMC5025882.
- Ciubotaru RM, Garcia-Aloy M, Masuero D, Franceschi P, Zulini L, Stefanini M, Oberhuber M, Robatscher P, Chitarrini G, Vrhovsek U. Semi-Targeted Profiling of the Lipidome Changes Induced by Erysiphe Necator in Disease-Resistant and Vitis vinifera L. Varieties. Int J Mol Sci. 2023 Feb 17;24(4):4072. doi: 10.3390/ijms24044072. PMID: 36835477; PMCID: PMC9958630.
- Watters DJ. Ascidian Toxins with Potential for Drug Development. Marine Drugs 2018, Vol 16, Page 162 [Internet]. 2018 May 13 [cited 2023 Jan 12];16(5):162. https://www.mdpi.com/1660-3397/16/5/162/htm
- Pascall M, Lee K. Food Regulations in the United States. Nutrition: An Approach to Good Health and Disease Management.]. 2016 [cited 2023 Jan 12];21.: https://books.google.com/books?hl=en&lr=&id=udrnDQAAQBAJ&oi=fnd&pg=PA21&dq=studies+have+demonstrated+that+many+polyketides,+whose+backbones+are+often+frequently+changed+by+glycosylation+or+oxidation,+e.g,+erythromycins,+tetracyclines,+avermectins,+are+commonly+utilized+anti-microbial,+anti-parasitic+and+anti-cancer+and+antitumor+&ots=Xz9T-tZ9Tl&sig=Cw8gfLR8x60ysDXmvNu3FwVILe4
- Seifbarghi S, Borhan MH, Wei Y, Coutu C, Robinson SJ, Hegedus DD. Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics. 2017 Mar 29;18(1):266. doi: 10.1186/s12864-017-3642-5. PMID: 28356071; PMCID: PMC5372324.
- Ranjan A, Westrick NM, Jain S, Piotrowski JS, Ranjan M, Kessens R, Stiegman L, Grau CR, Conley SP, Smith DL, Kabbage M. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol J. 2019 Aug;17(8):1567-1581. doi: 10.1111/pbi.13082. Epub 2019 Feb 11. PMID: 30672092; PMCID: PMC6662107.
- McCommis KS, Finck BN. Mitochondrial pyruvate transport: a historical perspective and future research directions. Biochem J. 2015 Mar 15;466(3):443-54. doi: 10.1042/BJ20141171. PMID: 25748677; PMCID: PMC4464838.
- Nikolaivits E, Pantelic B, Azeem M, Taxeidis G, Babu R, Topakas E, Brennan Fournet M, Nikodinovic-Runic J. Progressing Plastics Circularity: A Review of Mechano-Biocatalytic Approaches for Waste Plastic (Re)valorization. Front Bioeng Biotechnol. 2021 Jun 22;9:696040. doi: 10.3389/fbioe.2021.696040. PMID: 34239864; PMCID: PMC8260098.
- Chhatwal AR, Lomax HV, Blacker AJ, Williams JMJ, Marcé P. Direct synthesis of amides from nonactivated carboxylic acids using urea as nitrogen source and Mg(NO3)2 or imidazole as catalysts. Chem Sci. 2020 May 19;11(22):5808-5818. doi: 10.1039/d0sc01317j. PMID: 32832055; PMCID: PMC7416778.
- Teng Z, Yu Y, Zhu Z, Hong SB, Yang B, Zang Y. Melatonin elevated Sclerotinia sclerotiorum resistance via modulation of ATP and glucosinolate biosynthesis in Brassica rapa ssp. pekinensis. J Proteomics. 2021 Jul 15;243:104264. doi: 10.1016/j.jprot.2021.104264. Epub 2021 May 14. PMID: 33992838.
- Ranjan A, Westrick NM, Jain S, Piotrowski JS, Ranjan M, Kessens R, Stiegman L, Grau CR, Conley SP, Smith DL, Kabbage M. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol J. 2019 Aug;17(8):1567-1581. doi: 10.1111/pbi.13082. Epub 2019 Feb 11. PMID: 30672092; PMCID: PMC6662107.
- Lecompte F, Abro MA, Nicot PC. Can plant sugars mediate the effect of nitrogen fertilisation on lettuce susceptibility to two necrotrophic pathogens: Botrytis cinerea and Sclerotinia sclerotiorum? Plant Soil. 2013; Aug 11;369(1–2):387–401.
- Covington BC, McLean JA, Bachmann BO. Comparative mass spectrometry-based metabolomics strategies for the investigation of microbial secondary metabolites. Nat Prod Rep. 2017 Jan 4;34(1):6-24. doi: 10.1039/c6np00048g. PMID: 27604382; PMCID: PMC5214543.
- Chamberlain SA, Bronstein JL, Rudgers JA. How context dependent are species interactions? Ecol Lett. 2014 Jul;17(7):881-90. doi: 10.1111/ele.12279. Epub 2014 Apr 16. PMID: 24735225.
- Wiegand S, Jogler M, Boedeker C, Pinto D, Vollmers J, Rivas-Marín E, Kohn T, Peeters SH, Heuer A, Rast P, Oberbeckmann S, Bunk B, Jeske O, Meyerdierks A, Storesund JE, Kallscheuer N, Lücker S, Lage OM, Pohl T, Merkel BJ, Hornburger P, Müller RW, Brümmer F, Labrenz M, Spormann AM, Op den Camp HJM, Overmann J, Amann R, Jetten MSM, Mascher T, Medema MH, Devos DP, Kaster AK, Øvreås L, Rohde M, Galperin MY, Jogler C. Cultivation and functional characterization of 79 planctomycetes uncovers their unique biology. Nat Microbiol. 2020 Jan;5(1):126-140. doi: 10.1038/s41564-019-0588-1. Epub 2019 Nov 18. PMID: 31740763; PMCID: PMC7286433.
- Robison FM, Turner MF, Jahn CE, Schwartz HF, Prenni JE, Brick MA, Heuberger AL. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum. Plant Cell Environ. 2018 Sep;41(9):2141-2154. doi: 10.1111/pce.13176. Epub 2018 Mar 30. PMID: 29476531.
- Pandit MA, Kumar J, Gulati S, Bhandari N, Mehta P, Katyal R, Rawat CD, Mishra V, Kaur J. Major Biological Control Strategies for Plant Pathogens. Pathogens. 2022 Feb 19;11(2):273. doi: 10.3390/pathogens11020273. PMID: 35215215; PMCID: PMC8879208.