More Information
Submitted: October 07, 2022 | Approved: October 18, 2022 | Published: October 19, 2022
How to cite this article: Tall H, Tékété C, Comte A, Noba K, Hutin M, et al. Characterization of senegalese races of Xanthomonas oryzae PV. oryzae to identify resistance genes to use. J Plant Sci Phytopathol. 2022; 6: 135-145.
DOI: 10.29328/journal.jpsp.1001087
Copyright License: © 2022 Tall H, et al. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Keywords: Xanthomonas oryzae PV; Oryzae; Races; IRBB; Rice; Senegal
Characterization of senegalese races of Xanthomonas oryzae PV. oryzae to identify resistance genes to use
Tall H1*, Tékété C2, Comte A3, Noba K4, Hutin M3, Szurek B3, Verdier V3 and Cunnac S3
1Senegalese Institute of Agricultural Research (ISRA), Senegal
2University of Bamako, Faculty of Science and Technology, Mali
3PHIM, IRD, Cirad, University Montpellier, Montpellier, France
4Cheikh Anta Diop University of Dakar (UCAD), Botany and Biodiversity Laboratory, Senegal
*Address for Correspondence: Hamidou Tall, Senegalese Institute of Agricultural Research (ISRA), Senegal, Email: [email protected]
Bacterial blight (BB), is a disease caused by Xanthomonas oryzae PV. oryzae (Xoo), was first reported in Senegal by Trinh in 1980. BB represents a severe threat to rice cultivation in West Africa. Characterizing the pathotypic diversity of bacterial populations is a key to the management of pathogen-resistant varieties. Pathogenicity tests show that all strains are virulent on the susceptible rice variety Azucena, and interact differentially with twelve near-isogenic rice lines, each carrying a single resistance gene. On this rice panel, six races were identified, two of which were previously reported in Mali (A3) and Burkina Faso (A1). Four races (S2, S4, S5, and S6) are described for the first time in Africa. Races A1, isolated in Ndiaye and Ndioum areas is the most prevalent in Senegal. The Xa1 gene controls 100% of the isolates tested and xa5 controls all isolates except S4 strains. The geographical distribution of Xoo races is contrasted. Four races are detected in the North and two in the South East of the country. Race S4 can be a major risk to rice cultivation because strains from this race are the most virulent and can only be controlled by Xa1. To identify local sources of resistance, we screened Xoo strains representative of the various races on twenty-three rice varieties grown by farmers in Senegal. Four rice varieties namely Sahel210, Sangangbye, Dansna2, and Sahel305 effectively control all the isolates tested. Our characterization of the first collection of Senegalese Xoo strains provided insight into the races present in the country and identified sources of resistance in local rice varieties. This information will help design effective breeding programs for resistance to bacterial leaf blight in Senegal.
Rice is the second largest contributor to global human calorie intake [1] and its production continues to increase with the world’s population. Over the past 20 years, West Africa has experienced a large surge in rice consumption. Rice is a massively cultivated cereal and is the number one cereal plant of agronomic interest in Senegal in terms of production. Its production has more than quadrupled, from 231,805 tons in 2003 to 1,011,269 tons in 2018. But this production does not meet Senegal’s rice needs, which are estimated at 1,600,000 tons [2]. Although local production has progressed, through the expansion of planted areas and improvement of yields, Africa’s rice production remains insufficient to cover the rising needs that are driven by demographic growth, the increase in per capita consumption, and urbanization [3]. Bacterial blight (BB), caused by Xanthomonas oryzae PV. oryzae (Xoo), is one of the most widely distributed and devastating bacterial diseases on rice [4,5]. Xoo is a major constraint of rice production in almost all the paddy-growing regions [6]. Yield losses due to this disease generally vary from 20% to 30%, but sometimes these may go up to 50% [7,8]. The incidence of bacterial leaf blight was found to be aggravated by high dosages of nitrogen [9]. In addition to reducing yield, BB may also affect grain quality by interfering with maturation [7]. Although reported in Senegal as early as the 1980s [10], BB survey and sampling campaigns did not begin until 2014 and as a result, no rice breeding program for BB resistance has so far been developed in Senegal [11].
Xoo is a vascular pathogen that causes golden brown lesions along the leaf veins. Wounds and hydathodes are the main entry sites for Xoo. It multiplies in the epitome and subsequently enters into the xylem vessels where it starts active multiplication, leading to leaf wilting [12,13]. To colonize its host, Xoo has evolved a highly conserved type III secretion system (T3SS) that injects type III effectors into the plant cell. Type III effectors interfere with immune responses or facilitate nutritional or virulence processes to the benefit of the pathogen [14]. The Transcription Activator-Like Effectors (TALEs) type III effector family is composed of DNA-binding virulence proteins that modulate the expression of target genes. Some TALEs act as major virulence factors that are essential for BB to occur and that target rice susceptibility genes [15]. Type III secretion substrates also act as avirulence factors in incompatible interactions on resistant hosts and can trigger a defense reaction called the hypersensitive response (HR) 16]. The nine conserved groups of TALE identified in the genome of African Xoo [17,18] appear to be genetically distant from Asian ones [19].
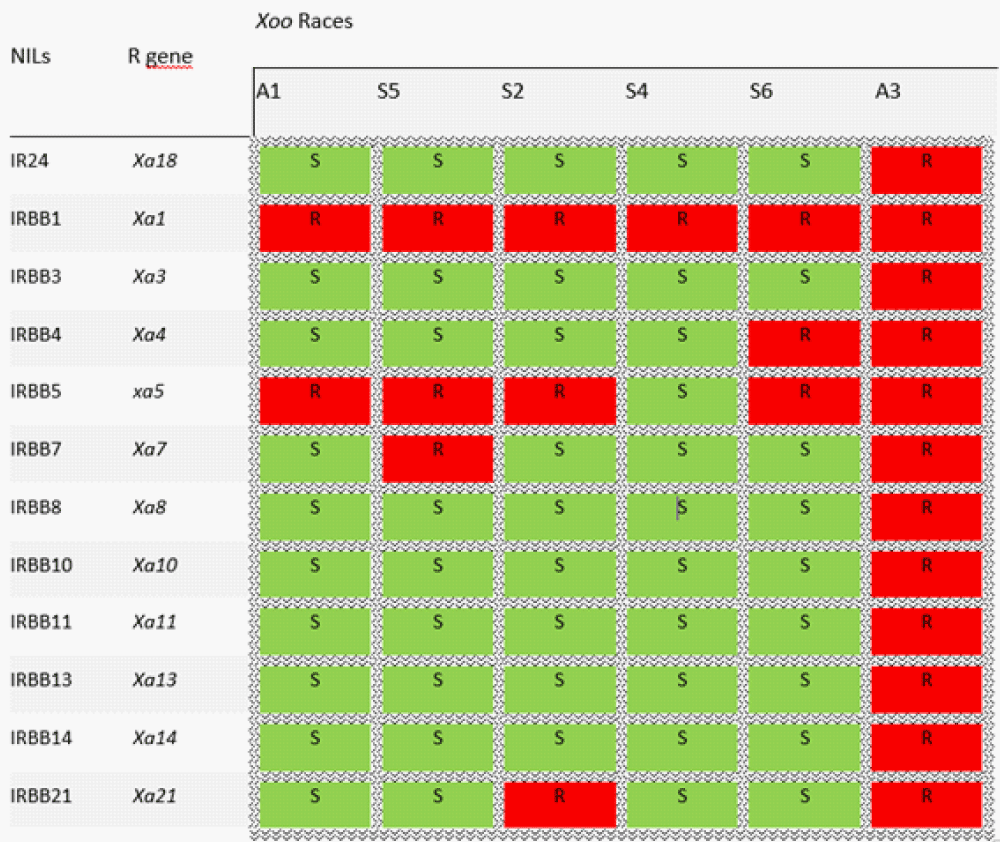
BB control methods include agronomic practices, and chemical and biological control, all of which have proven ineffective in the long term [20-22]. Breeding and deployment of resistant cultivars carrying major resistance genes have been the most effective approach for BB management [23-27] and several resistance genes have been incorporated into elite rice cultivars [23]. However, the deployment of uniform, monogenic resistances over vast areas have decreased their effectiveness, because it provided a selection pressure for the emergence of virulent pathotypes [28,29]. It is therefore critical to monitor pathogen populations to detect new isolates that are able to overcome the resistance genes deployed in the field. A set of Near-Isogenic rice Lines (NILs), each carrying a single resistance gene and the recurrent parent IR24 are commonly used to characterize the race structure of the pathogen [30]. A Xoo race is a group of strains sharing the same virulence profile on a panel of these rice NILS. In West-Africa, three races have been identified with race A3 reported in Mali, race A1 and A2 in Burkina Faso, and A1 also reported in Niger [19]. In Mali, using twelve NILs, Tekete, et al. recently identified six new races (A4 to A9) in addition to race A3 [31].
Although BB was first reported in Senegal in the 1980s [10] and confirmed only recently [11], there has been no rigorous assessment of the incidence of this disease and no pathogen strain has been isolated. So far, in the absence of information on the pathogen and its impact on rice, these bacteria have not been considered threats to rice production. Thus no rice selection program for BB resistance has been developed in Senegal. The aim of this study was to update and expand our knowledge of Xoo in Senegal. To do this, surveys were conducted in the main rice-growing regions of the country and the race structure of the isolated strains of Xoo was determined. Finally, we assessed the susceptibility to BB of a set of Senegalese rice varieties to identify local sources of resistance to this disease.
Surveys and isolation of X. oryzae PV. oryzae
Surveys and isolation of X. oryzae PV. oryzae Surveys for rice BB disease were performed in 2015 and 2016 in major rice production areas in Senegal. Field visits were conducted from September to October of each year during the rainy season when symptoms were visible. Leaves with BB-like symptoms (translucent, yellow blight, sometimes with visible bacterial exudates on the leaf surface) were collected from Oryza sativa cultivars and wild rice species (Oryza barthii). The isolation of the bacteria was carried out from a fragment of leaf showing symptoms of BB. A 5 cm fragment of infected leaf including a seemingly healthy part was disinfected successively in 75% ethanol and 0.1% bleach, rinsed two times in sterilized distilled water, and dried on blotting paper. The leaf fragment was then cut into small pieces in a 2 ml Eppendorf tube containing 2 marbles. The tube was immersed in liquid nitrogen for 10 minutes by cryogrinding as described by Schoutten c. and al, 1986 and ground by shaking vigorously and 1 ml of sterilized distilled water is added to the leaf powder. The resulting solution was left to rest for at least 30 minutes and stirred periodically. A 50 μl aliquot of this solution is spread over the PSA culture medium (10 g Peptone, 10 g Sucrose, 16 g Agar and 1 g glutamic acid) supplemented with antibiotics (cephalexin 1ml (40 mg/ml), Kasugamicin 1 ml (20 mg/ml) and Actidione 1ml (50 mg/ml), for 1 l of culture medium). The Petri dishes were incubated in an inverted position for 72 hours at 28 °C. Colonies similar in appearance to that of Xanthomonas oryzae (light yellow, circular, mucous membrane, convex, bright, smooth) were purified on antibiotic-free PSA. The strains were stored at -80 °C in tubes containing liquid nutrient media (PSA 70% and glycerol 30%).
Multiplex PCR
All X. oryzae PV. oryzae strains were identified using a multiplex PCR specific for X. oryzae pathovars. Four primer pairs specific to X. oryzae (Xo3756F; CATCGTTAGGACTGCCAGAAG and Xo3756R: GTGAGAACCACCGCCATCT), to X. oryzae PV. oryzae (Xoo80F: GCCGCTAGGAATGAGCAAT and Xoo80R: GCGTCCTCGTCTAAGCGATA) to X. oryzae PV oryzicola (i.e. Xoc3866F: ATCTCCCAGCATGTTGATCG and Xoc3866R: GCGTTCAATCTCCTCCATGT) and to universal for bacteria (Univ-0008-a-S-19F: GAGTTTGATCCTGGCTCAG [32]. For DNA extraction, bacteria were grown on a PSA medium at 28 °C. DNA extraction was carried out from a 48-hour bacterial culture in a solid PSA medium using the Gram-negative Bacteria-specific Promega Genomic DNA Purification Kit System (following the recommendations of the supplier). PCR with X. oryzae-specific multiplex primer was performed as previously described by Lang, et al. [32]. All isolates were tested at least twice.
Pathogenicity assays
The evaluation of the pathogenicity of Xoo strains was carried out on the susceptible rice variety Azucena. The characterization of Xoo isolates in virulence groups (races) was carried out on a set of twelve Near Isogenic Lines (IRBB) each carrying a specific resistance gene (Xa) (Table 1) and the recurrent parent IR24 using a simplified scale developed by IRRI [33]. The seeds of the IRBB varieties were obtained from IRRI as part of an application by the Senegalese Institute for Agricultural Research (ISRA) for the search for a source of resistance against Xoo isolates. The resistance of twenty-three varieties of rice grown in Senegal was also assessed. The seeds were obtained from the selection department of the Saint Louis ISRA Center. Pathogenicity tests were conducted in a greenhouse at IRD, Montpellier, France (temperature and relative humidity: day 28 °C and 80%; at night 25 °C and 60%) and on field plots (temperature and relative humidity: maximum 35 °C and 90% minimum 22 °C and 60%) at the ISRA/ Kolda experimentation station, City, Senegal.
Rice seeds were sown in terrines filled with a “Siffy” substrate. For experiments with plants growing in the field, a Split-plot experimental setup was adopted. It included 3 repetitions with a large plot the variety within which the strains will be arranged randomly. Two factors were studied, a ’variety’ factor with 12 modalities in race test and 23 in screening, and a ‘strain’ factor with 46 modalities. In field assays, each plot consisted of a line of 4 meters long and 2 meters wide with a 20-centimeter gap between the lines and between the pockets. A total of 200 plants per plot fertilized with NPK 15-15-15 due to 200 kg per hectare, and nitrogen fertilizer with 46% nitrogen due to 150 kg per hectare (in two fractions; half at the tillering stage and another half at the mounting stage). The inoculum was prepared with sterilized distilled water and bacterial colonies from a culture on a solid PSA medium incubated at 28 °C for 24h. Bacterial suspensions were adjusted to an optical density at 600nm of 0.2 (~108 bacteria/mL) using a spectrophotometer. Forty-five days-old plants were inoculated by leaf-clipping using the Kauffman method [34]. A total of 9 plants and 3 leaves per plant were inoculated for each treatment.
Disease assessment
Lesion lengths from the cut leaf tip were measured in centimeters 15 days after inoculation [29]. Disease reactions were categorized according to lesion lengths, where plants with an average value of less than 5 cm were designated as resistant, and sensitive (S) otherwise [29,33,35].
Statistical analysis
Lesion length (LL) data for each rice line - Xoo isolates combination were used to evaluate the responses of rice lines to BB. A two-factor variance analysis (ANOVA) was performed following Fisher’s Least Significant Difference (LSD). The means were separated by Fisher’s least important tests (p < 0.05). Isolates were grouped into races or pathotypes when presenting the same virulence profile on NILs or Senegalese varieties respectively. This data was analyzed with the R statistical software to perform Hierarchical Clustering (HC). Races are described according to their virulence profiles R (resistance) and/or S (sensitive) on IRBB varieties. The Complex Heatmap package was used to build and visualize the various heatmaps generated in this study [36].
A survey of Xoo isolates in the main regions of rice production in Senegal
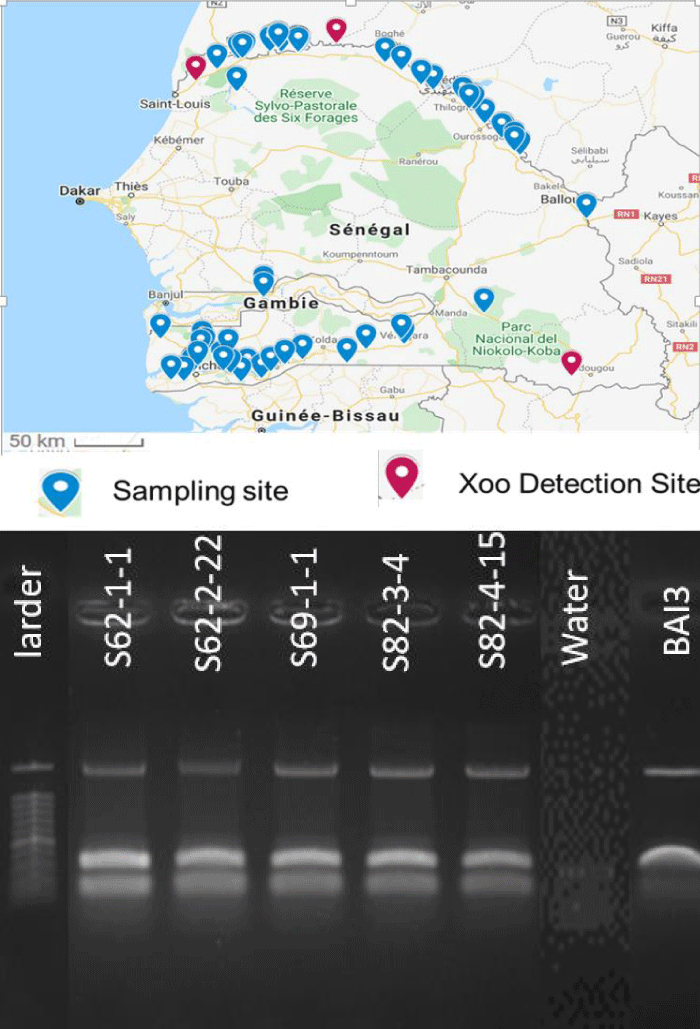
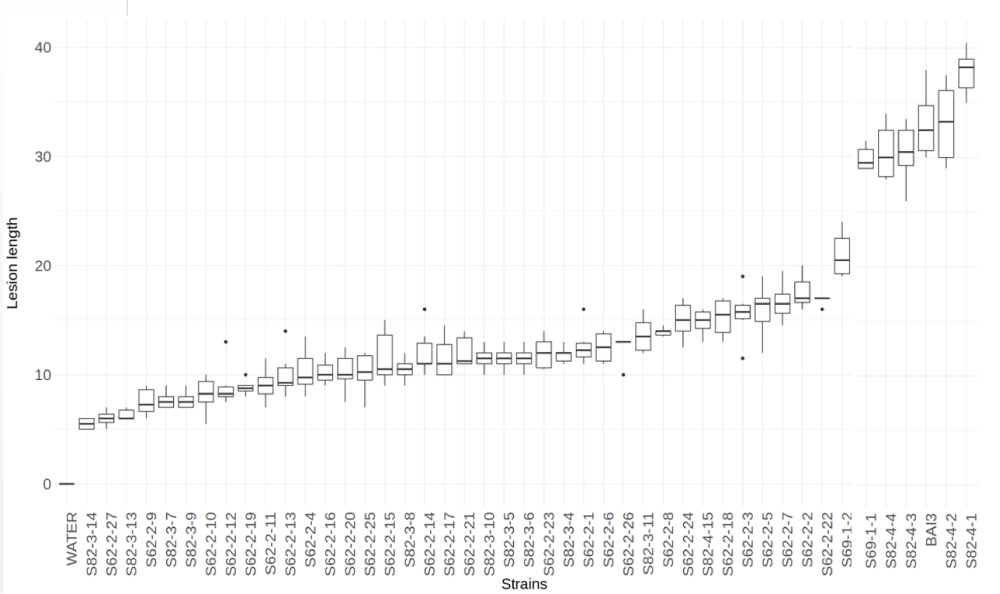
Field surveys were conducted during the rainy season in the main areas of rice production corresponding to the southern and northern regions of Senegal and part of the Tambacounda and Kedougou regions (Figure 1a,b). The distance between 2 consecutive sampling points (10 km to 20 km) depended on both rice field size and the diversity of the cultivated varieties. Samples were taken from infected rice leaves displaying symptoms of BB in the fields. For each sample, passport data was recorded (date and place of collection, code number, GPS coordinates, name of the site, and the name of the variety on which the sample was taken...). From September to October of the year 2015 and 2016, a total of 53 sites were visited and 103 samples were randomly collected from rice plants showing symptoms of BB. One leaf per sample was processed for bacterial isolation. In total, forty-four Xoo isolates forming pale yellow colonies on the PSA medium, were isolated and confirmed by multiplex PCR. Overall, Xoo strains were isolated from three sites (Ndiaye, Ndioum, and Bandafassi), corresponding to ten leaf samples that were collected on the most cultivated varieties in Senegal or on the wild species Oryza barthii (Table 1). To confirm their pathogenicity, all isolates were tested on the susceptible rice variety Azucena using a leaf-clipping assay. Typical symptoms of BB were visible for all isolates 14 days after inoculation. Lesion lengths caused by the isolates varied from 6 cm to 37 cm (Figure 2).
| Table 1:List of Xanthomonas oryzae pv. oryzae strains characterized in this study. | |||||||||||
| Strain | Other names | Origin | Region | Site | Year | Host | MLSA | Virulence e | Race | Reference | |
| S62-2-1 | CIX2353 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-2 | CIX2354 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-3 | CIX2355 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-4 | CIX2356 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S2 | This study | |
| S62-2-5 | CIX2357 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S2 | This study | |
| S62-2-6 | CIX2358 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-7 | CIX2359 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-8 | CIX2360 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-9 | CIX2361 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-10 | CIX2362 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-11 | CIX2363 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-12 | CIX2364 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-13 | CIX2365 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-14 | CIX2366 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-15 | CIX2367 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-16 | CIX2368 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-17 | CIX2369 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-18 | CIX2370 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-19 | CIX2371 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-20 | CIX2372 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S5 | This study | |
| S62-2-21 | CIX2373 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-22 | CIX2374 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S5 | This study | |
| S62-2-23 | CIX2375 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-24 | CIX2376 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S5 | This study | |
| S62-2-25 | CIX2377 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S62-2-26 | CIX2378 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | S6 | This study | |
| S62-2-27 | CIX2379 | Senegal | Saint Louis | Ndiaye | 2016 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S69-1-1 | CIX2951 | Senegal | Saint Louis | Ndioum | 2017 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S69-1-2 | CIX2952 | Senegal | Saint Louis | Ndioum | 2017 | Oryza sativa | Xoo Africa | 5 | A1 | This study | |
| S82-3-4 | CIX2964 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-5 | CIX2965 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-6 | CIX2966 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-7 | CIX2967 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-8 | CIX2968 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-9 | CIX2969 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-10 | CIX2970 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-11 | CIX2971 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-13 | CIX2972 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-3-14 | CIX2973 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | A3 | This study | |
| S82-4-1 | CIX2974 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | S4 | This study | |
| S82-4-2 | CIX2975 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | S4 | This study | |
| S82-4-3 | CIX2976 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | S4 | This study | |
| S82-4-4 | CIX2977 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | S4 | This study | |
| S82-4-15 | CIX2978 | Senegal | Kédougou | Bandafassi | 2017 | Oryza bartei | Xoo Africa | 5 | S4 | This study | |
| Xanthomonas oryzae PV. oryzae (used as controls) | |||||||||||
| MAI1 | CFBP7325, LMG25985 | Mali | O.Niger | Niono | 2003 | O.sativa | Xoo Africa | na | A3 | Gonzalez et al, 2007 | |
| BAI3 | CFBP7321, LMG25983 | Burkina | East Center | Bagre | 2003 | O.sativa | Xoo Africa | na | A1 | Gonzalez et al, 2007 | |
Figure 1: a: Sampling map and sites were Xanthomonas oryzae PV. oryzae strains were isolated. Diseased leaf samples were collected in the rice-growing fields of Senegal river valley in the North, Tambacounda, and Kedougou regions in the East and South East, and the Casamance region. The location of the surveyed sites is represented as blue pins on the map. Sites, where Xoo was isolated, are indicated on the map with red pins. b: Agarose gel of five Senegalese Xoo strains and two controls (BAI3 and water).
Figure 2: Xanthomonas oryzae PV. oryzae pathogenicity assay on the rice variety Azucena.
This indicates that all the isolates tested are virulent on Azucena, some of them being able to kill the plants after two months. One-way analysis of variance on lesion length revealed that there is a significant difference among isolates (p < 0.001). Compared to Xoo BAI3, the symptoms caused by strains from Senegal vary. Because some are comparable to BAI3 while others have caused shorter but significant lesions on Azucena. Altogether, our data (PCR and pathogenicity test) confirmed that the 44 isolates are indeed virulent Xoo bacteria (Table 1). Also, the complete genome of strain S62-2-22/ CIX2374 (Table 1) was sequenced by Perez-Quintero, A.L. in 2019 and reflected at National Center for Biotechnology Information ( NCBI) under the reference NZ_CP036377.1. The comparison of the genome of this strain to those of PXO99 (a strain of the Philippines) and BAI3 (a strain of Burkina Faso) confirms that the strains isolated are 100% African Xoo.
Box plots of the lesion length data by Xoo isolate. The X-axis indicates the Xoo isolates tested. Y-axis indicates the lesion length in cm measured 15 days after inoculation using the leaf-clipping method. The boxes represent the quartiles while the inside bars correspond to the median. Data from at least three independent replicates of the experiment with each 9 replicate lesion length values per treatment. The dots represent values that are out of the 5% and 95% quartiles delineated by the whiskers. Xoo BAI3 was used as a control.
Identification of Senegalese Xoo races from 2015 to 2016 surveys
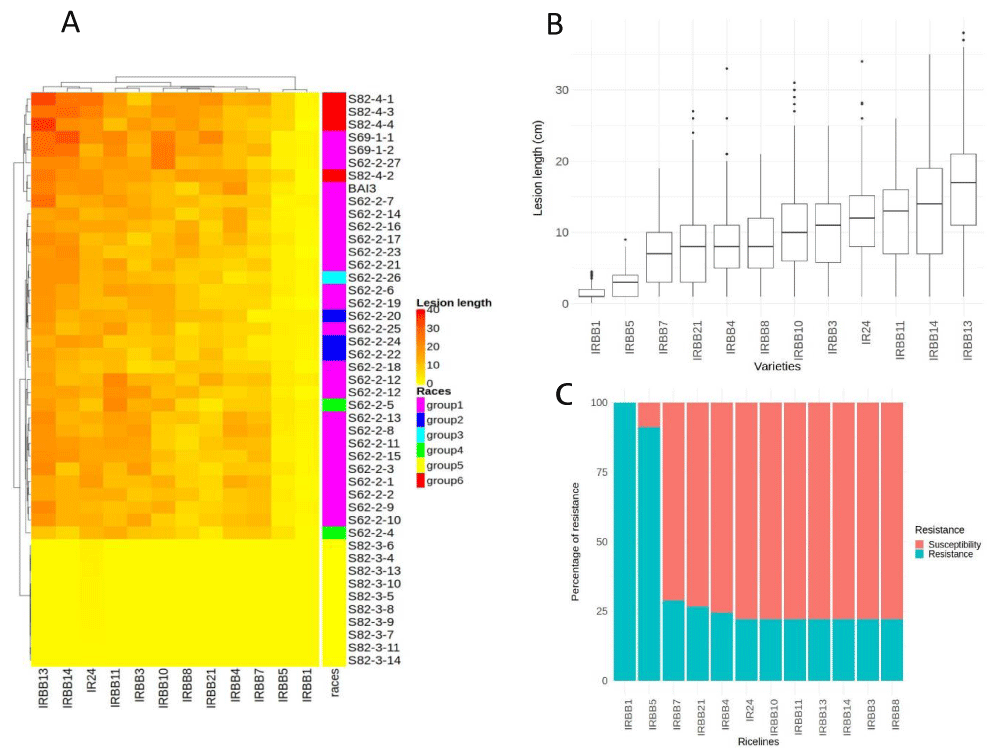
The efficacy of Xa resistance genes was evaluated based on leaf clipping assay data of the 44 Xoo strains inoculated on 12 IRBB lines each containing a single R gene. IRBB lines exhibited different reaction profiles upon challenge with the 44 Xoo isolates (Figure 3). All IRBB lines were susceptible to one or more of our Senegalese Xoo strains except IRBB1 which was scored as resistant to all strains. Lesion length varied from 1 cm to 38 cm (Figure 3B). IRBB lines have different levels of resistance (Figure 3C). Similarly, variance analysis (ANOVA) shows that there is a significant difference between the IRBBs and the isolates tested p < 0.001). IR24, the recurrent parent, is sensitive to 34 of the 46 isolates tested. Most IRBBs were sensitive to the majority of isolates (Figure 3A). Hierarchical clustering of lesion length data for this experiment shows that IRBB1 is resistant to all Xoo strains and that IRBB5 is sensitive to only one group of isolates (Figure 3C, Table 2). Thus, Xa1 and xa5 provide broad resistance.
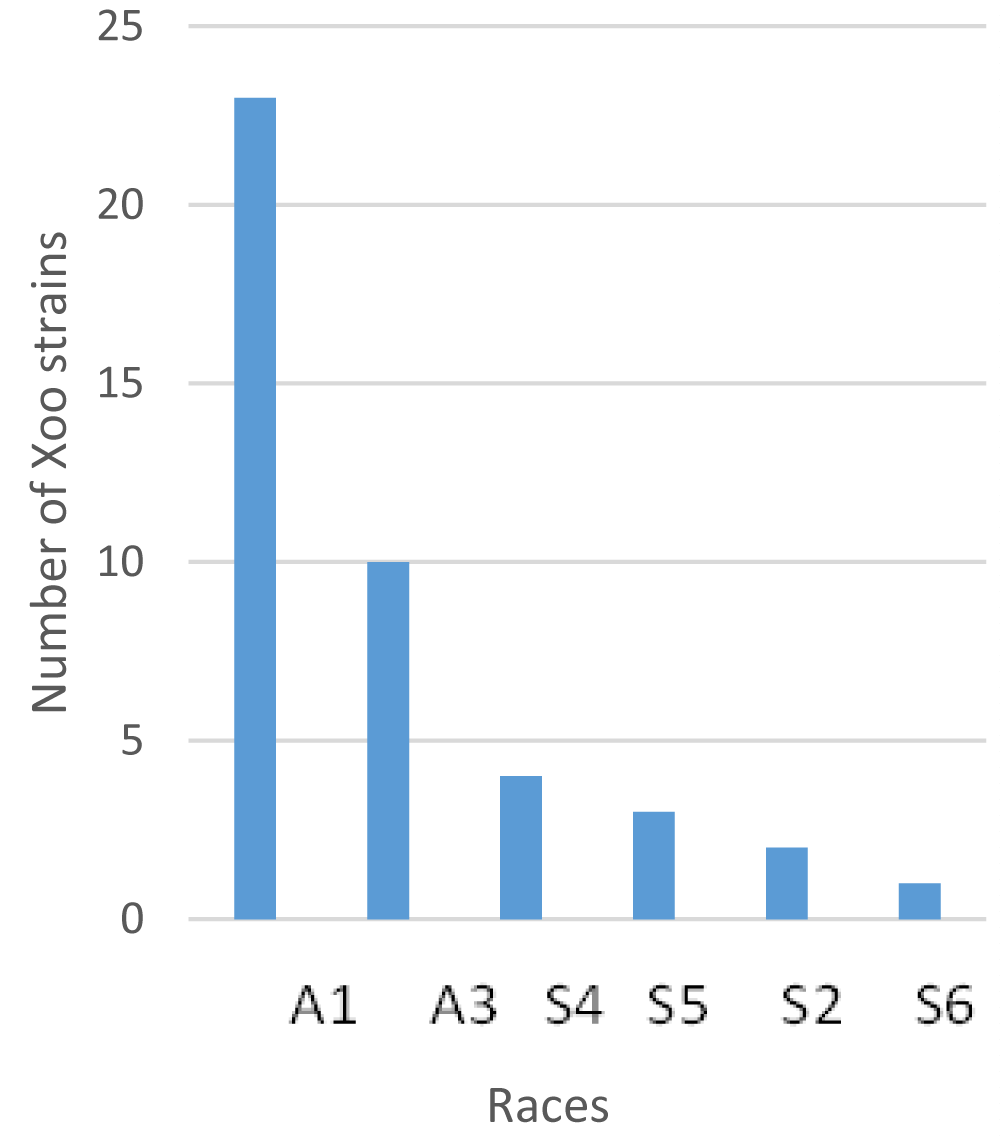
The same dataset was analyzed for the definition of Xoo races based on interactions between isolates and IRBB lines. The forty-four Xoo strain tested were classified into six races (named A1, S2, A3, S4, S5, and S6) following a categorical analysis of lesion length associated with each combination of IRBB line and Xoo isolate (Figure 3A, Table 2). The A1 race is the most represented with almost 53.5% of the isolates tested and the S6 race, is the least represented with only 2.3% of the isolates (Table 1, Figure 4). The A3 race is avirulent on all IRBB lines and the recurrent parent IR24, while the S4 race is virulent on all IRBB lines except IRBB1 (Table 2, Figure 3C). Of the list of assayed resistance genes, Xa1 is the only one that can control all the identified Xoo races (Table 2). The recessive gene xa5 also controls all races except the S4 race (Table 2).
Figure 3: IRBB Xa resistance efficacity again forty-four Xoo strains.
Figure 4: Number of strains collected in Senegal within the different races identified.
Table 2: Virulence of Senegalese Xanthomonas oryzae PV. oryzae strain against NILs varieties containing a single Xa gene in IR24 genetic background: 44 Xoo strains (41 strains from this study and 4 reference strains) cluster into six races. Each race is characterized by its virulence profile on NILs. The ability of Xa genes to control different races is displayed. R = resistant (Lesion Length ≤ 5 cm) and S = susceptible (Lesions length > 5 cm).
The geographical distribution of the different races of Xoo in Senegal is shown in Table 1. Isolates from Ndiaye (Figure 1) cover 4 out of 6 races and are the most diverse with regard to their virulence profile on the IRBB lines (Table 1, Figure 4). It should also be noted that no single region houses all described races. The A3 and S4 races are specifically present in the East of the country while the A1, S2, S5, and S6 races are confined to the Senegal River Valley in the North (Table 1, Figure 4). The A1 race is the most predominant with 23 out of 43 strains of Xoo and is present in two localities in the North while in the South East it is the A3 race (Table 3, Figure 4).
| Table 3: Geographical distribution and race classification of Xoo isolated from 2015 to 2016 in Senegal. The number of strains belonging to each race is indicated. | |||||||
| Locality | Xoo Races | Total strains | |||||
| A1 | S5 | S2 | S4 | S6 | A3 | ||
| Ndiaye | 21 | 3 | 2 | 0 | 1 | 0 | 27 |
| Ndioum | 2 | 0 | 0 | 0 | 0 | 0 | 2 |
| Bandafassi | 0 | 0 | 0 | 4 | 0 | 10 | 14 |
| Total strain | 23 | 3 | 2 | 4 | 1 | 10 | 43 |
Virulence profile clustering of Xoo isolates on a set of twelve IRBB lines. On the heatmap, each interaction (IRBB lines x Xoo), (i.e individual cell colors) codes for the corresponding mean lesion length across three independent replicates experiment with every 9 values. Races are color-coded in the colored bar on the right. Isolates were also clustered according to their quantitative virulence profiles using a hierarchical clustering approach based on pairwise Euclidian distances as displayed in the tree on the left. The susceptibility profiles of the NILs were also clustered using the same method as displayed on the hierarchical tree above the heatmap. (B) The X-axis indicates the IRBB lines. Y-axis indicates the lesion lengths in cm observed 15 days after inoculation by leaf clipping. The displayed data (box plots) corresponds to lesion lengths of three repetitions per treatment (combination IRBBs x isolates). (C) Relative frequency of resistance (R in blue) or susceptibility (S in red) reactions against Senegalese isolates for individual IRBB lines. Reactions with an average lesion length < 5 cm were considered resistant, and ≥ 5 cm were considered susceptible.
Screening 23 rice varieties for resistance to BB
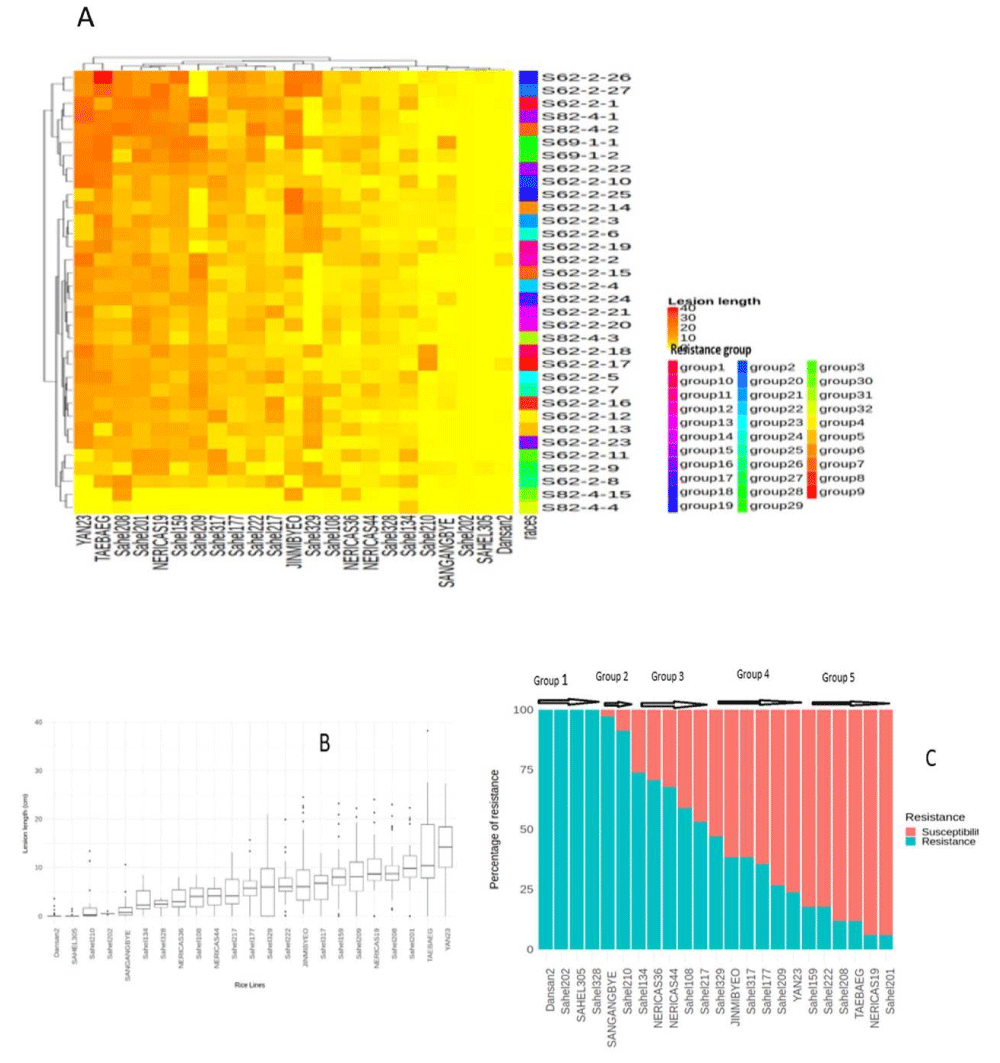
Given the limited number of R genes in the tested IRBB panel capable of controlling the different Xoo races encountered in Senegal (Table 2), we felt it necessary to identify additional resistance sources from 23 domestically grown varieties (Table 4) in order to diversify and adapt sources of resistance to BB. These varieties were released relatively recently compared to older varieties (such as DJ and ITA) and are most often cultivated in irrigated and low-land systems by farmers. Our Senegalese variety panel was challenged as before with 35 Xoo strains representing all previously identified races except race A3 strains because it is not virulent on any of the varieties except the Azucena variety. ANOVA indicates that there is a significant difference in virulence between the isolates (p < 0,001). Different responses were also noted among the varieties (Figure 5). There is a large variation in the value of the median length of lesions per variety and these values range between 0 cm and 46.1 cm (Figure 5B). Average lesion length values per treatment were converted to a categorical scale and classified as ‘susceptible’ if the average length was equal or superior to 5 cm or ‘resistant’ otherwise. Similar to what is generally done for the classification of strains into races, we classified strains in the same pathotype if their categorical interaction profile on the Senegalese varieties were identical (Figure 5A). The hierarchical dendrogram of the average lesion length data induced by 35 strains on the 23 varieties was cut into 32 Clusters which correspond to different resistance profiles (Figure 5A). Nineteen varieties are found to be susceptible to at least one race of identified Xoo strain and four varieties (Dansan2; Sahel202, Sahel305, and Sahel328) are resistant to all strains tested (Figure 5C). Based on the percentage of strains these varieties are resistant to, they can be subdivided into five different groups (Figure 5C). The first group includes varieties resistant to 100% of the strains (4 varieties); the second is composed of varieties resistant to more than 75% of the strains (2 varieties); the third group corresponds to those varieties that are resistant to between 75% and 50% of strains (4 varieties); the fourth group includes those varieties that are resistant to between 50% and 25% of the strains (5 varieties). Finally, varieties classified in the fifth group are resistant to less than 25% of the strains (Figure 5C).
| Table 4: List of Senegalese varieties used in the resistance screen. | |||||||||
| Ecology | Variety name | Parents | species | origin | Cycle (days) | BB resistance | Creation year | Diffusion Year | Potential Yield (T/H) |
| Irrigated | Sahel 210 | na | O. Sativa (Indica) | Latin America | sensitive | na | 2007 | 12 | |
| Irrigated | Sahel 217 | Sahel 201/4456 | O. sativa (Indica) | AfricaRice | 109-129 | Tolerant | 1997 | 2009 | 13 |
| Irrigated | Sahel 209 | TSY/MOROBERKAN//ITA306 | O. sativa (Indica) | IITA/Nigeria | 126-140 | Tolerant | na | 2007 | 12 |
| Irrigated | Sahel 208 | ITA 212/UPL RI 7 | O. sativa (Indica) | IITA/Nigeria | 125-145 | Tolerant | na | 2007 | 12 |
| Irrigated | Sahel 201 | IR 2071-586/BG 400-1 | O. sativa (Indica) | Sri Lanka | 121-142 | Tolerant | na | 1994 | 10 |
| Irrigated | Sahel 222 | Sahel 201/4456 | O. sativa (Indica) | AfricaRice | 103-129 | Tolerant | 1997 | 2009 | 13 |
| Irrigated | Sahel 305 | IR 64/4456 | O. sativa (Indica) | AfricaRice | 101-124 | Tolerant | 1995 | 2009 | 10 |
| Irrigated | Sahel 317 | 4456/32 Xuan 5C | O. sativa (Indica) | AfricaRice | 97-122 | Tolerant | 1995 | 2009 | 12 |
| Irrigated | Sahel 328 | IR 31851-96-2-3-2-1(Sahel 134)/IR 66231-37-1-2 | O. sativa (Indica) | AfricaRice | 87-116 | Tolerant | 1997 | 2009 | 10 |
| Irrigated | Sahel 329 | Jaya / Basmati 370 | O. sativa (Indica) | AfricaRice | 87-116 | Tolerant | 1997 | 2009 | 10 |
| Irrigated | Sahel 108 | IR 30 (BHP)/BABAWE//IR 36 | O. sativa (Indica) | IRRI Philippines | 110-117 | Tolerant | na | 1994 | 7,5 |
| Irrigated | Sahel 134 | IR 1791-5-4-3-3/IR 9129-209-2-2-2-1 | O. sativa (Indica) | IRRI Philippines | 110-131 | Tolerant | na | 2007 | 10 |
| Irrigated | Sahel 159 | IR 13240-108-2-2-3/IR 9129-209-2-2-2-1 | O. sativa (Indica) | IRRI Philippines | 110-130 | Tolerant | na | 2007 | 10 |
| Irrigated | Sahel 177 | IR 31851-96-2-3-2-1(Sahel 134)/IR 66231-37-1-2 | O. sativa (Indica) | AfricaRice | 110-122 | Tolerant | 1998 | 2009 | 10 |
| Irrigated | NERICA S 19 | Tog 5681/2*IR 64//IR 31785 | Oryza sativa x Oryza glaberrima | AfricaRice | 105-131 | na | 1997 | 2009 | 11 |
| 16 | NERICA S 44 | IR 64//Tog 5681/4*IR 64 | Oryza sativa x Oryza glaberrima | AfricaRice | 100-122 | na | 1998 | 2009 | 12 |
| 17 | NERICA S 36 | Tog 5681/2*IR 1529//IR 1529 | Oryza sativa x Oryza glaberrima | AfricaRice | 101-120 | na | 1998 | 2009 | 11 |
| 18 | Dansan2 | na | O. sativa (Indica) | na | na | Tolerant | na | na | na |
| 19 | MYRLANG 23 | IR 1317-316-5-1/IR 24 | Tongil (japonica/indica) | South Korea | na | na | na | 2017 | 13,5 |
| 20 | TAEBAEGRYEO | IR 24*2/IR 747-B2-6-3 | Tongil (japonica/indica) | South Korea | na | na | na | 2017 | 12,5 |
| 21 | SAMGAMBYEO | na | Tongil (japonica/indica) | South Korea | na | Tolerant | na | na | 12,5 |
| 22 | JINMIBYEO K9) | na | Tongil (japonica/indica) | South Korea | na | na | na | na | - |
| Irrigated | Sahel 202 | TOX 494-3696/TOX 711/BG 6812 | O. sativa (Indica) | Nigeria | 117-139 | na | 1994 | 11 | |
| Each variety is listed by pedigree, species and subspecies, origin (country or institution), cycle, year of diffusion, and potential yield. IRRI: International Rice Research Institute, Philippines, Africa Rice: Africa Rice Center, Senegal; IITA: International Institute of Tropical Agriculture, Ibadan, Niger, na: data not available | |||||||||
Figure 5: Senegalese rice varieties level resistance to 35 Xoo strains.
(A) Aggressiveness profile of isolates on the rice varieties tested. Heatmap display of the aggregated lesion length data by isolate and by variety. For each interaction (rice varieties Xoo isolates), individual cell colors code for the corresponding mean lesion length across three independent replicates of the experiment with each six replicates. Strains are color-coded colored bar on the right. (B) Efficiency of rice varieties against the 35 isolates tested. X-axis indicates the rice varieties tested; Y-axis indicates the lesions lengths in cm observed 15 days after inoculation by leaf clipping. Overall lesion length data per rice varieties. The displayed data (box plots) corresponds to lesion lengths of three repetitions per treatment (rice varieties 35 isolates). (C) Frequency of resistance phenotypes among the rice varieties. Lesion lengths < 5 cm are considered as resistant (R in blue), and ≥ 5 cm susceptible (S in pink).
Sampling for plant leaves with BB symptoms has been conducted at several sites but the isolation of bacteria was only successful in three sites despite the high number of sites surveyed. This may be due to the isolation protocol which has some limitations in extracting bacteria from the diseased tissues. For this reason it would be interesting to perform the multiplex PCR test directly on the leaf lysates since we cannot exclude that the selected portion no longer contains living Xoo. Xoo isolates from a collection of rice leaf samples from different regions of Senegal were characterized, revealing the presence of six races in the set of 44 identified Xoo strains. These 6 races are the first Xoo characterized in Senegal following the confirmation of the presence of the pathogen by Tall, et al. [11]. Xa1 stands out as the most effective R-gene capable of controlling all six Senegalese races. Furthermore, xa5 also shows broad efficiency but remains ineffective against strains of the S4 race. Importantly, four rice varieties grown in Senegal, namely Dansan2, Sahel202, Sahel305 and Sahel328, were found to be resistant to all Senegalese Xoo strains tested.
The first characterization of Xoo races from Senegal points to Xa1 and xa5 as efficient R-genes against this pathogen population.
One of the major objectives of this work was to study the pathotypic diversity of Xoo strains isolated from different geographic areas in Senegal. In this study, all 12 IRBB nearly-isogenic lines showed different levels of resistance. Phenotypical diversity of Xoo exists throughout Senegal. The virulence profiles of Xoo collected on improved and wild rice were similar, suggesting that host diversity does not affect pathogenicity on the set of rice genotypes tested here (Table 1, Figure 2). Several authors have reported that the diversity of hosts does not affect the pathogenicity of Xoo populations [37]. However, other authors argue that host diversity has an effect on the pathogenicity of Xanthomonas oryzae pv. oryzae [38]. Our virulence analysis of 44 strains collected in 3 Senegalese rice-producing areas indicates that Xoo strains can be classified into 4 Senegalese-specific races (S2, S4, S5 and S6) that are different from those previously reported in West Africa [19,30,31,39]. However, our results confirmed the presence of race A3 that has been described in Mali and race A1 described in Burkina Faso [19,30,31,40]. None of the isolated Xoo strains from Senegal were able to overcome the Xa1 resistance gene. Similarly, the xa5 gene controls all Xoo races except S4 (Figure 3A and 3C). Thus, the xa5 and Xa1 genes have good potential for breeding BB resistant rice varieties in Senegal. Two Xoo races (A1 and S4) showed similar reactions on the 12 NILs, except on IRBB5, harboring the R xa5 gene. The S4 race was the only one virulent on IRBB5. Several studies have shown that the degree of resistance conferred by xa5 varies for Xoo strains from different regions of West Africa [31,41,42]. A majority of Xoo races (83%) was virulent on IRBB3, IRBB8, IRBB10, IRBB11, IRBB13 and IRBB14 suggesting that the genes Xa3, xa8, Xa10, Xa11, Xa13 and Xa14 cannot provide rice resistance against most Xoo strains in Senegal. The Xa21 gene controls only 2 races out of the 6 identified races. This indicates that Xa21 action spectrum is limited against Senegalese Xoo races. This results is in line with previous reports [31,40]. Altogether, these studies conclude on a limited practical value of Xa21 to globally control African Xoo races. This contrasts with other studies that have reported the broad efficiency of the Xa21 gene against Asian races of Xoo [43,44]. In conclusion, we believe that pyramiding Xa1 and xa5 R-genes would be most effective in providing broad and potentially durable BB resistance to Senegalese rice varieties. These two R-genes could be transferred to susceptible elite cultivars via marker-assisted selection. On the other hand, it will be difficult to understand the evolutionary trajectories of Senegalese Xoo because there are no isolates prior to H. Tall sampling in 2019 [11].
We also examined if the geographical distribution of Senegalese Xoo races displayed any detectable pattern. When cross-referencing race test results with geographic sampling data (Table 1), we observed that races A1, S2, S5 and S6 are exclusively detected in the Senegal River Valley in the North of the country, while the A3 and S4 races were only found in the South-East. We therefore conclude that Senegalese races segregate along a North/South pattern. The probable hypothesis at this origin of differentiation may be due to the fact that two different agro-ecologies can be distinguished with intensive irrigated system in the North and low-land rainfed system in the South-East with specific rice varieties adapted to each ecology. Our results highlight the importance of continuously monitoring Senegalese Xoo population using IRBB lines as a discrimination tool.
Varieties Sahel202, Sahel305, Sahel328 and Dansan2 possess broad resistance against Senegalese Xoo
The rice varieties used in this screening study are listed in Table 2. A total of 46 strains were tested, including the BAI3 strain [19] as a positive control and distilled sterilize water as a negative control for leaf inoculations in all trials. In the set of 23 rice varieties widely cultivated in Senegal and screened for bacterial blight (BB) resistance, accessions have various levels of resistance to BB (Figure 5C). Six of them are highly and broadly resistant to all tested strains, with Sahel202, Sahel305, Sahel328 and Dansan2 being the most promising. These 4 varieties may carry new genes for BB resistance with broad resistance spectrum against Xoo populations in Senegal. Similar approaches have been carried out in West Africa, notably in Mali and Burkina Faso [31,45] which also identified domestic varieties with good potential as BB resistance donors in breeding programs. In this respect, the characterization of the genetic determinants of the resistance of Sahel202, Sahel305, Sahel328 and Dansan2 needs further investigation.
In conclusion, this study on Senegalese Xoo virulence diversity and host resistance profiles is a pioneering work in the country and parallels recent progresses on BB pathology in the sub-region of West Africa. We studied isolates collected over two years to investigate the nature and diversity of pathogenic races, their geographic distribution and the R-genes resistance needed to control BB. The collected Xoo strains were subdivided into 6 races displaying a geographic distribution pattern with 4 races in the North and 2 in the South-East of Senegal. Compared to other studies in West Africa, the composition of Senegalese Xoo races indicates the presence of races already described in Mali and Burkina Faso, and 4 new races described for the first time in Africa. The Xa1 and xa5 R-genes are the most promising candidate genes for a broad control of Xoo races detected to date. The study also identified several varieties grown in Senegal that are highly resistant to different races detected in the country. However, further analysis is needed if we want to understand and identify the R-genes that are involved in this resistance. The transfer of this resistance to other high-yielding varieties grown in Senegal is expected to significantly improve rice productivity and BB control.
We thank Doctors Allan Salabsabin, IRRI, Philippines and Omar Ndaw Faye, ISRA, Senegal for providing respectively the seeds of near-isogenic lines and Senegalese rice varieties; Doctor B. Maneh, AfricaRice, Senegal for his help given in sampling for BB rice leaves.
Author contributions
H.T., K.N. and V.V. designed the research; H.T., K.N.; S.C. and V.V. performed the research; H.T., S.C., A.C and V.V analysed the data; H.T., K.N., S.C., B.S, A.C., M.H. and V.V wrote the manuscript.
- Chauhan Bhagirath S, Jabran K, Mahajan G. Rice Production Worldwide. Springer. 2017.
- DAPSA n.d. Agricultural Survey Results (Provisional) | Department of Analysis, Forecasting and Agricultural Statistics - DAPSA. http://www.dapsa.gouv.sn/content/statistiques-agricoles. Consulté le 29 octobre 2019. http://www.dapsa.gouv.sn/content/r%C3%A9sultats-enqu%C3%AAte-agricole-2018-pr%C3%A9visionnels. 2018.
- Patricio Mendez del V, Jean-Martin B. Rice in West Africa: dynamics, policies and perspectives. Cahiers Agricultures 2013;22 (5): 336-344 (1). https://doi.org/10.1684/agr.2013.0657.
- Mew TW. Current Status and Future Prospects of Research on Bacterial Blight of Rice. Annual Review of Phytopathology 1987;25(1): 359‑82. https://doi.org/10.1146/annurev.py.25.090187.002043.
- Niño-Liu DO, Ronald PC, Bogdanove AJ. Xanthomonas oryzae pathovars: model pathogens of a model crop. Mol Plant Pathol. 2006 Sep;7(5):303-24. doi: 10.1111/j.1364-3703.2006.00344.x. PMID: 20507449.
- George ML, Bustamam M, Cruz WT, Leach JE, Nelson RJ. Movement of Xanthomonas oryzae pv. oryzae in Southeast Asia Detected Using PCR-Based DNA Fingerprinting. Phytopathology. 1997 Mar;87(3):302-9. doi: 10.1094/PHYTO.1997.87.3.302. PMID: 18945173.
- Shu Huang Ou. Commonwealth Mycological Institute Great Britain. Rice Diseases. IRRI. 1985.
- Sukhwinder-Singh, Sodhi M, Vikal Y, George MLC, Bala GS, Mangat GS, Garg M, Sidhu JS, Dhaliwal HSM. DNA Fingerprinting and Virulence Analysis of Xanthomonas oryzae Pv. oryzae Isolates from Punjab, Northern India. Euphytica 2003; 130 (1): 107‑15. https://doi.org/10.1023/A:1022329024651.
- Awoderv VA, Bangura N, John VT. Incidence, distribution and severity of bacterial diseases on rice in West Africa. Tropical Pest Management 37 (2): 113‑17. https://doi.org/10.1080/09670879109371553. 1991.
- Trinh TT. New Rice Diseases and Insects in the Senegal River Basin in 1978/79. International Rice Commission Newsletter. 1980;29 (2). https://www.cabdirect.org/cabdirect/abstract/19810581551.
- Hamidou T, Tekete C, Noba K, ousmane K, Sebastien C, Mathilde H, Boris S, Valérie Verdier. Confirmation report of Bacterial blight caused by Xanthomonas oryzae pv. oryzae on rice in Senegal . Plant Disease, novembre. https://doi.org/10.1094/PDIS-07-19-1464-PDN. 2019.
- Köplin R, Arnold W, Hötte B, Simon R, Wang G, Pühler A. Genetics of Xanthan Production in Xanthomonas Campestris: The XanA and XanB Genes Are Involved in UDP-Glucose and GDP-Mannose Biosynthesis. Journal of Bacteriology. 1992;174 (1): 191‑99. https://doi.org/10.1128/jb.174.1.191-199.1992.
- Ray SK, Rajeshwari R, Sonti RV. Mutants of Xanthomonas oryzae pv. oryzae deficient in general secretory pathway are virulence deficient and unable to secrete xylanase. Mol Plant Microbe Interact. 2000 Apr;13(4):394-401. doi: 10.1094/MPMI.2000.13.4.394. PMID: 10755302.
- Lindgren PB. The role of hrp genes during plant-bacterial interactions. Annu Rev Phytopathol. 1997;35:129-52. doi: 10.1146/annurev.phyto.35.1.129. PMID: 15012518.
- White FF, Potnis N, Jones JB, Koebnik R. The type III effectors of Xanthomonas. Mol Plant Pathol. 2009 Nov;10(6):749-66. doi: 10.1111/j.1364-3703.2009.00590.x. PMID: 19849782; PMCID: PMC6640274.
- Alfano JR, Collmer A. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385-414. doi: 10.1146/annurev.phyto.42.040103.110731. PMID: 15283671.
- Tran TT, Pérez-Quintero AL, Wonni I, Carpenter SCD, Yu Y, Wang L, Leach JE, Verdier V, Cunnac S, Bogdanove AJ, Koebnik R, Hutin M, Szurek B. Functional analysis of African Xanthomonas oryzae pv. oryzae TALomes reveals a new susceptibility gene in bacterial leaf blight of rice. PLoS Pathog. 2018 Jun 4;14(6):e1007092. doi: 10.1371/journal.ppat.1007092. PMID: 29864161; PMCID: PMC6037387.
- Doucouré H, Pérez-Quintero AL, Reshetnyak G, Tekete C, Auguy F, Thomas E, Koebnik R, Szurek B, Koita O, Verdier V, Cunnac S. Functional and Genome Sequence-Driven Characterization of tal Effector Gene Repertoires Reveals Novel Variants With Altered Specificities in Closely Related Malian Xanthomonas oryzae pv. oryzae Strains. Front Microbiol. 2018 Aug 6;9:1657. doi: 10.3389/fmicb.2018.01657. PMID: 30127769; PMCID: PMC6088199.
- Gonzalez C, Szurek B, Manceau C, Mathieu T, Séré Y, Verdier V. Molecular and pathotypic characterization of new Xanthomonas oryzae strains from West Africa. Mol Plant Microbe Interact. 2007 May;20(5):534-46. doi: 10.1094/MPMI-20-5-0534. PMID: 17506331.
- Devadath S, Padmanabhan SY. Approaches to Control of Bacterial Blight and Streak Diseases of Rice in India. Bulletin of the Indian Phytopathological Society, no No. 1970;6: 5‑12.
- Ganeshan G, Manoj Kumar A. Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. Journal of Plant Interactions 2005;1 (3): 123‑34. https://doi.org/10.1080/17429140600907043.
- Ahmed HU, Finckh MR, Alfonso RF, Mundt CC. Epidemiological effect of gene deployment strategies on bacterial blight of rice. Phytopathology. 1997 Jan;87(1):66-70. doi: 10.1094/PHYTO.1997.87.1.66. PMID: 18945155.
- Tsugufumi O, Yamamoto T, Gurdev S. Khush, Twng-Wah Mew. Breeding of Near-Isogenic Line of Rice With Single Gene for Resistance to Bacterial Blight Pathogen (Xanthomonas compestris pv. oryzae) . Journal of Breeding Studies 1991; 41 (3): 523‑29. https://doi.org/10.1270/jsbbs1951.41.523.
- Huang N, Angeles ER, Domingo J, Magpantay G, Singh S, Zhang G, Kumaravadivel N, Bennett J, Khush GS. Pyramiding of Bacterial Blight Resistance Genes in Rice: Marker-Assisted Selection Using RFLP and PCR . Theoretical and Applied Genetics 1997;95 (3): 313‑20. https://doi.org/10.1007/s001220050565.
- KINOSHITA T. Report of the committee on gene symbolization, nomenclature and linkage groups. Rice Genet. Newslett. 1990;7: 16‑50.
- Rao KK, Lakshminarasu M, Jena KK. DNA markers and marker-assisted breeding for durable resistance to bacterial blight disease in rice. Biotechnol Adv. 2002 Apr;20(1):33-47. doi: 10.1016/s0734-9750(02)00002-2. PMID: 14538061.
- Cheema KK, Grewal NK, Vikal Y, Sharma R, Lore JS, Das A, Bhatia D, Mahajan R, Gupta V, Bharaj TS, Singh K. A novel bacterial blight resistance gene from Oryza nivara mapped to 38 kb region on chromosome 4L and transferred to Oryza sativa L. Genet Res (Camb). 2008 Oct;90(5):397-407. doi: 10.1017/S0016672308009786. PMID: 19061530.
- Mew TW. International Rice Research Institute, C. M. Vera Cruz, et E. S. Medalla. 1992. Changes in Race Frequency of Xanthomonas oryzae Pv. oryzae in Response to Rice Cultivars Planted in the Philippines. Plant Disease (USA). http://agris.fao.org/agris-search/search.do?recordID=US9317148.
- Adhikari, Tika B, Mew TW, Leach JE. Genotypic and Pathotypic Diversity in Xanthomonas oryzae pv. oryzae in Nepal . PhytopathologyTM 1999;89 (8): 687‑94. https://doi.org/10.1094/PHYTO.1999.89.8.687.
- Lindsay T, Koebnik R, Valerie V, Jan E. Leach. The Genomics of Xanthomonas oryzae . In Genomics of Plant-Associated Bacteria, édité par Dennis C. Gross, Ann Lichens-Park, Chittaranjan Kole, 2014 ;127‑50. Berlin, Heidelberg: Springer. https://doi.org/10.1007/978-3-642-55378-3_6.
- Tekete C, Cunnac S, Doucouré H, Dembele M, Keita I, Sarra S, Dagno K, Koita O, Verdier V. Characterization of New Races of Xanthomonas oryzae pv. oryzae in Mali Informs Resistance Gene Deployment. Phytopathology. 2020 Feb;110(2):267-277. doi: 10.1094/PHYTO-02-19-0070-R. Epub 2019 Dec 19. PMID: 31464159.
- Lang JM, Hamilton JP, Diaz MGQ, Van Sluys MA, Burgos MRG, Vera Cruz CM, Buell CR, Tisserat NA, Leach JE. Genomics-Based Diagnostic Marker Development for Xanthomonas oryzae pv. oryzae and X. oryzae pv. oryzicola. Plant Dis. 2010 Mar;94(3):311-319. doi: 10.1094/PDIS-94-3-0311. PMID: 30754246.
- Quibod IL, Perez-Quintero A, Booher NJ, Dossa GS, Grande G, Szurek B, Vera Cruz C, Bogdanove AJ, Oliva R. Effector Diversification Contributes to Xanthomonas oryzae pv. oryzae Phenotypic Adaptation in a Semi-Isolated Environment. Sci Rep. 2016 Sep 26;6:34137. doi: 10.1038/srep34137. PMID: 27667260; PMCID: PMC5035989.
- Kauffman HE. An improved technique for evaluat-ing resistance of rice varieties to Xanthomonas oryzae. Plant Dis. Rep 1973;57: 537‑41.
- Debabrata N, Bose LK, Singh UD, Sanjay S, Parsuram Nayak sd. Measurement of Genetic Diversity of Virulence in Populations of Xanthomonas oryzae , 14.
- Leland W. Michael Friendly. 2009. The History of the Cluster Heat Map. The American Statistician 63 (2): 179‑84. https://doi.org/10.1198/tas.2009.0033.
- Adhikari TB, Mew TW, Teng PS. Phenotypic Diversity of Xanthomonas oryzae Pv. oryzae in Nepal. Plant Disease (USA). http://agris.fao.org/agris-search/search.do?recordID=US9434820. 1994.
- Takahito N, Chengyun Li, Jiarui Li, Hirokazu Ochiai, Kazuo Ise, Hisatoshi Kaku. Pathogenic Diversity of Xanthomonas oryzae pv. oryzae Strains from Yunnan Province, China . Japan Agricultural Research Quarterly: JARQ 2001;35 (2): 97‑103. https://doi.org/10.6090/jarq.35.2.
- Hinda d. Création de résistance à large spectre contre la bactériose foliaire du riz au Mali . Thesis, Montpellier. http://www.theses.fr/2017MONTT100. 2017.
- Wonni Issa. Les bactérioses du riz dues à Xanthomonas oryzae au Burkina Faso: Diversité identification de sources de résistance adaptées. PhD Thesis, Montpellier 2. 2013.
- Yu Y, Streubel J, Balzergue S, Champion A, Boch J, Koebnik R, Feng J, Verdier V, Szurek B. Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Mol Plant Microbe Interact. 2011 Sep;24(9):1102-13. doi: 10.1094/MPMI-11-10-0254. PMID: 21679014.
- Djedatin G, Ndjiondjop MN, Mathieu T, Cruz CMV, Sanni A, Ghesquière A, Verdier V. Evaluation of African Cultivated Rice Oryza glaberrima for Resistance to Bacterial Blight. Plant Dis. 2011 Apr;95(4):441-447. doi: 10.1094/PDIS-08-10-0558. PMID: 30743359.
- Wen-Ling D, Heng-An Lin, Yu-Cyuan S, Kuo CH, Tzeng JU, Li-yu D. Liu, Shu-Tzu Huang, Chi-Ming Huang, Chia-Lin Chung. Genotypic and Pathotypic Diversity of Xanthomonas oryzae pv. oryzae Strains in Taiwan . Journal of Phytopathology 2016;164 (10): 745‑59. https://doi.org/10.1111/jph.12495.
- Wang GL, Song WY, Ruan DL, Sideris S, Ronald PC. The cloned gene, Xa21, confers resistance to multiple Xanthomonas oryzae pv. oryzae isolates in transgenic plants. Mol Plant Microbe Interact. 1996 Dec;9(9):850-5. doi: 10.1094/mpmi-9-0850. PMID: 8969533.
- Issa W. Evaluation of Elite Rice Varieties Unmasks New Sources of Bacterial Blight and Leaf Streak Resistance for Africa. Rice Research: Open Access. 2016; 4 (1). https://doi.org/10.4172/2375-4338.1000162.